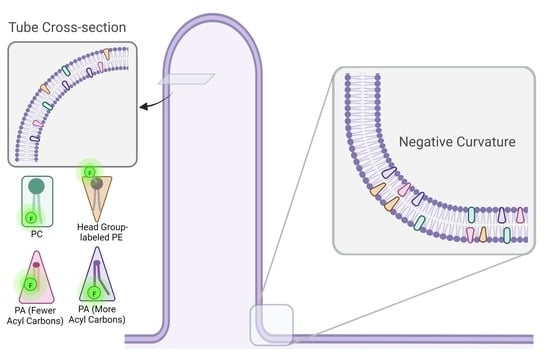
Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Liposome and STuBs Assembly
2.2. Fluorescence Recovery after Photobleaching (FRAP)
2.3. Total Internal Reflection Fluorescence Microscopy
2.4. Fluorimetry
3. Results
3.1. The Formation of STuBs Depends on NaCl Concentration
3.2. PA and DPPE Form More Tubules Than PC
3.3. Longer Fatty Acid Chains Support Tubule Formation
3.4. Dithionite Quenching of NBD Reveals PA Localization to Negative Curvature
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hodgkin, M.N.; Pettitt, T.R.; Martin, A.; Michell, R.H.; Pemberton, A.J.; Wakelam, M.J.O. Diacylglycerols and Phosphatidates: Which Molecular Species Are Intracellular Messengers? Trends Biochem. Sci. 1998, 23, 200–204. [Google Scholar] [CrossRef]
- Thakur, R.; Naik, A.; Panda, A.; Raghu, P. Regulation of Membrane Turnover by Phosphatidic Acid: Cellular Functions and Disease Implications. Front Cell Dev. Biol. 2019, 7, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Devaiah, S.P.; Zhang, W.; Welti, R. Signaling Functions of Phosphatidic Acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef] [PubMed]
- Rohrbough, J.; Broadie, K. Lipid Regulation of the Synaptic Vesicle Cycle. Nat. Rev. Neurosci. 2005, 6, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Kooijman, E.E.; Chupin, V.; Fuller, N.L.; Kozlov, M.M.; de Kruijff, B.; Burger, K.N.J.; Rand, P.R. Spontaneous Curvature of Phosphatidic Acid and Lysophosphatidic Acid. Biochemistry 2005, 44, 2097–2102. [Google Scholar] [CrossRef] [PubMed]
- Cheney, P.P.; Weisgerber, A.W.; Feuerbach, A.M.; Knowles, M.K. Single Lipid Molecule Dynamics on Supported Lipid Bilayers with Membrane Curvature. Membranes 2017, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Cooke, I.R.; Deserno, M. Coupling between Lipid Shape and Membrane Curvature. Biophys. J. 2006, 91, 487–495. [Google Scholar] [CrossRef] [Green Version]
- Callan-Jones, A.; Sorre, B.; Bassereau, P. Curvature-Driven Lipid Sorting in Biomembranes. Cold Spring Harb. Perspect Biol. 2011, 3. [Google Scholar] [CrossRef] [Green Version]
- Kooijman, E.E.; Chupin, V.; de Kruijff, B.; Burger, K.N.J. Modulation of Membrane Curvature by Phosphatidic Acid and Lysophosphatidic Acid. Traffic 2003, 4, 162–174. [Google Scholar] [CrossRef]
- Hatzakis, N.S.; Bhatia, V.K.; Larsen, J.; Madsen, K.L.; Bolinger, P.Y.; Kunding, A.H.; Castillo, J.; Gether, U.; Hedegård, P.; Stamou, D. How Curved Membranes Recruit Amphipathic Helices and Protein Anchoring Motifs. Nat. Chem. Biol. 2009, 5, 835–841. [Google Scholar] [CrossRef]
- Bhatia, V.K.; Hatzakis, N.S.; Stamou, D. A Unifying Mechanism Accounts for Sensing of Membrane Curvature by BAR Domains, Amphipathic Helices and Membrane-Anchored Proteins. Semin. Cell Dev. Biol. 2010, 21, 381–390. [Google Scholar] [CrossRef]
- Nuscher, B.; Kamp, F.; Mehnert, T.; Odoy, S.; Haass, C.; Kahle, P.J.; Beyer, K. α-Synuclein Has a High Affinity for Packing Defects in a Bilayer Membrane. J. Biol. Chem. 2004, 279, 21966–21975. [Google Scholar] [CrossRef] [Green Version]
- Parthasarathy, R.; Yu, C.; Groves, J.T. Curvature-Modulated Phase Separation in Lipid Bilayer Membranes. Langmuir 2006, 22, 5095–5099. [Google Scholar] [CrossRef]
- Pucadyil, T.J.; Schmid, S.L. Real-Time Visualization of Dynamin-Catalyzed Membrane Fission and Vesicle Release. Cell 2008, 135, 1263–1275. [Google Scholar] [CrossRef] [Green Version]
- Hanson, L.; Lin, Z.C.; Xie, C.; Cui, Y.; Cui, B. Characterization of the Cell–Nanopillar Interface by Transmission Electron Microscopy. Nano Lett. 2012, 12, 5815–5820. [Google Scholar] [CrossRef]
- Black, J.C.; Cheney, P.P.; Campbell, T.; Knowles, M.K. Membrane Curvature Based Lipid Sorting Using a Nanoparticle Patterned Substrate. Soft Matter 2014, 10, 2016–2023. [Google Scholar] [CrossRef] [Green Version]
- Kabbani, A.M.; Woodward, X.; Kelly, C. v Revealing the Effects of Nanoscale Membrane Curvature on Lipid Mobility. Membranes 2017, 7, 60. [Google Scholar] [CrossRef] [Green Version]
- Kamal, M.M.; Mills, D.; Grzybek, M.; Howard, J. Measurement of the Membrane Curvature Preference of Phospholipids Reveals Only Weak Coupling between Lipid Shape and Leaflet Curvature. Proc. Natl. Acad. Sci. USA 2009, 106, 22245–22250. [Google Scholar] [CrossRef] [Green Version]
- Schenk, N.A.; Dahl, P.J.; Hanna, M.G.; Audhya, A.; Tall, G.G.; Knight, J.D.; Anantharam, A. A Simple Supported Tubulated Bilayer System for Evaluating Protein-Mediated Membrane Remodeling. Chem. Phys Lipids 2018, 215, 18–28. [Google Scholar] [CrossRef]
- Welti, R.; Li, W.; Li, M.; Sang, Y.; Biesiada, H.; Zhou, H.-E.; Rajashekar, C.B.; Williams, T.D.; Wang, X. Profiling Membrane Lipids in Plant Stress Responses. J. Biol. Chem. 2002, 277, 31994–32002. [Google Scholar] [CrossRef]
- Zech, T.; Ejsing, C.S.; Gaus, K.; de Wet, B.; Shevchenko, A.; Simons, K.; Harder, T. Accumulation of Raft Lipids in T-Cell Plasma Membrane Domains Engaged in TCR Signalling. EMBO J. 2009, 28, 466–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelstein, A.; Amodaj, N.; Hoover, K.; Vale, R.; Stuurman, N. Computer Control of Microscopes Using ΜManager. Curr. Protoc. Mol. Biol. 2010, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barg, S.; Knowles, M.K.; Chen, X.; Midorikawa, M.; Almers, W. Syntaxin Clusters Assemble Reversibly at Sites of Secretory Granules in Live Cells. Proc. Natl. Acad. Sci. USA 2010, 107, 20804–20809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knowles, M.K.; Barg, S.; Wan, L.; Midorikawa, M.; Chen, X.; Almers, W. Single Secretory Granules of Live Cells Recruit Syntaxin-1 and Synaptosomal Associated Protein 25 (SNAP-25) in Large Copy Numbers. Proc. Natl. Acad. Sci. USA 2010, 107, 20810–20815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crocker, J.C.; Grier, D.G. Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interface Sci. 1996, 179, 298–310. [Google Scholar] [CrossRef] [Green Version]
- Blair, D.; Dufresne, E. The Matlab Particle Tracking Code Repository. 2005. Available online: https://site.physics.georgetown.edu/matlab/ (accessed on 10 June 2014).
- Alnaas, A.A.; Moon, C.L.; Alton, M.; Reed, S.M.; Knowles, M.K. Conformational Changes in C-Reactive Protein Affect Binding to Curved Membranes in a Lipid Bilayer Model of the Apoptotic Cell Surface. J. Phys. Chem. B 2017, 121, 2631–2639. [Google Scholar] [CrossRef]
- Diaz, A.J.; Albertorio, F.; Daniel, S.; Cremer, P.S. Double Cushions Preserve Transmembrane Protein Mobility in Supported Bilayer Systems. Langmuir 2008, 24, 6820–6826. [Google Scholar] [CrossRef] [Green Version]
- Karatekin, E.; di Giovanni, J.; Iborra, C.; Coleman, J.; O’Shaughnessy, B.; Seagar, M.; Rothman, J.E. A Fast, Single-Vesicle Fusion Assay Mimics Physiological SNARE Requirements. Proc. Natl. Acad. Sci. USA 2010, 107, 3517–3521. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Michelot, A.; Koskela, E.V.; Tkach, V.; Stamou, D.; Drubin, D.G.; Lappalainen, P. Membrane-Sculpting BAR Domains Generate Stable Lipid Microdomains. Cell Rep. 2013, 4, 1213–1223. [Google Scholar] [CrossRef] [Green Version]
- Angeletti, C.; Nichols, J.W. Dithionite Quenching Rate Measurement of the Inside−Outside Membrane Bilayer Distribution of 7-Nitrobenz-2-Oxa-1,3-Diazol-4-Yl-Labeled Phospholipids. Biochemistry 1998, 37, 15114–15119. [Google Scholar] [CrossRef]
- McIntyre, J.C.; Sleight, R.G. Fluorescence Assay for Phospholipid Membrane Asymmetry. Biochemistry 1991, 30, 11819–11827. [Google Scholar] [CrossRef]
- Volinsky, R.; Cwiklik, L.; Jurkiewicz, P.; Hof, M.; Jungwirth, P.; Kinnunen, P.K.J. Oxidized Phosphatidylcholines Facilitate Phospholipid Flip-Flop in Liposomes. Biophys. J. 2011, 101, 1376–1384. [Google Scholar] [CrossRef] [Green Version]
- Ladokhin, A.S.; Selsted, M.E.; White, S.H. Sizing Membrane Pores in Lipid Vesicles by Leakage of Co-Encapsulated Markers: Pore Formation by Melittin. Biophys. J. 1997, 72, 1762–1766. [Google Scholar] [CrossRef] [Green Version]
- Huster, D.; Müller, P.; Arnold, K.; Herrmann, A. Dynamics of Membrane Penetration of the Fluorescent 7-Nitrobenz-2-Oxa-1,3-Diazol-4-Yl (NBD) Group Attached to an Acyl Chain of Phosphatidylcholine. Biophys. J. 2001, 80, 822–831. [Google Scholar] [CrossRef] [Green Version]
- Loura, L.M.S.; Ramalho, J.P.P. Location and Dynamics of Acyl Chain NBD-Labeled Phosphatidylcholine (NBD-PC) in DPPC Bilayers. A Molecular Dynamics and Time-Resolved Fluorescence Anisotropy Study. Biochim. Biophys. Acta BBA Biomembr. 2007, 1768, 467–478. [Google Scholar] [CrossRef] [Green Version]
- Harlos, K.; Eibl, H. Hexagonal Phases in Phospholipids with Saturated Chains: Phosphatidylethanolamines and Phosphatidic Acids. Biochemistry 1981, 20, 2888–2892. [Google Scholar] [CrossRef]
- Verkleij, A.J.; de Maagd, R.; Leunissen-Bijvelt, J.; de Kruijff, B. Divalent Cations and Chlorpromazine Can Induce Non-Bilayer Structures in Phosphatidic Acid-Containing Model Membranes. Biochim. Biophys. Acta BBA Biomembr. 1982, 684, 255–262. [Google Scholar] [CrossRef]
- Balch, C.; Morris, R.; Brooks, E.; Sleight, R.G. The Use of N-(7-Nitrobenz-2-Oxa-1,3-Diazole-4-Yl)-Labeled Lipids in Determining Transmembrane Lipid Distribution. Chem. Phys. Lipids 1994, 70, 205–212. [Google Scholar] [CrossRef]
- Mills, J.K.; Needham, D. Lysolipid Incorporation in Dipalmitoylphosphatidylcholine Bilayer Membranes Enhances the Ion Permeability and Drug Release Rates at the Membrane Phase Transition. Biochim. Biophys. Acta BBA Biomembr. 2005, 1716, 77–96. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Soe, T.T.; Maxfield, F.R. Endocytic Sorting of Lipid Analogues Differing Solely in the Chemistry of Their Hydrophobic Tails. J. Cell Biol. 1999, 144, 1271–1284. [Google Scholar] [CrossRef]
- Cullis, P.R.; de Kruijff, B. Lipid Polymorphism and the Functional Roles of Lipids in Biological Membranes. Biochim. Biophys. Acta BBA Rev. Biomembr. 1979, 559, 399–420. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bills, B.L.; Knowles, M.K. Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes. Biomolecules 2022, 12, 1707. https://doi.org/10.3390/biom12111707
Bills BL, Knowles MK. Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes. Biomolecules. 2022; 12(11):1707. https://doi.org/10.3390/biom12111707
Chicago/Turabian StyleBills, Broderick L., and Michelle K. Knowles. 2022. "Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes" Biomolecules 12, no. 11: 1707. https://doi.org/10.3390/biom12111707
APA StyleBills, B. L., & Knowles, M. K. (2022). Phosphatidic Acid Accumulates at Areas of Curvature in Tubulated Lipid Bilayers and Liposomes. Biomolecules, 12(11), 1707. https://doi.org/10.3390/biom12111707