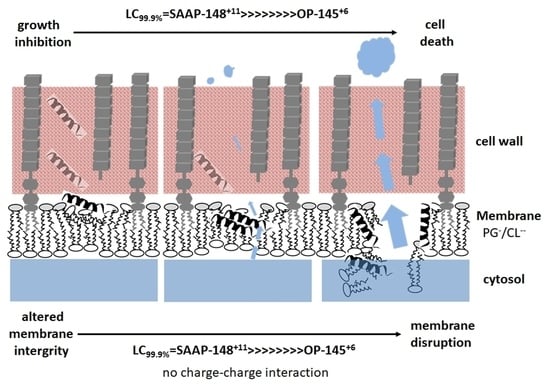
Membrane Activity of LL-37 Derived Antimicrobial Peptides against Enterococcus hirae: Superiority of SAAP-148 over OP-145
Abstract
:1. Introduction
2. Experimental Procedures
2.1. In Silico Evaluation of the Peptide’s Structural Properties
2.2. Materials
2.3. Methods
2.3.1. Growth Conditions of Enterococcus hirae
2.3.2. Preparation of Model Membranes
2.3.3. Antimicrobial Activity of OP-145 and SAAP-148
2.3.4. Depolarization of E. hirae Membrane and Fluorescence Microscopy
2.3.5. Membrane Permeabilization of E. hirae and Flow Cytometry
2.3.6. Staining of E. hirae Membranes and Fluorescence Microscopy
2.3.7. Permeability of Model Membrane and Fluorescence Spectroscopy
2.3.8. Phosphate Determination
2.3.9. Surface Charge Neutralization of Model Membranes and ζ-Potential Measurements
2.3.10. Thermotropic Phase Behavior of Model Membranes and Differential Scanning Calorimetry (DSC)
3. Results
3.1. OP-145 and SAAP-148 Kill E. hirae in a Concentration and Time-Dependent Manner
3.2. SAAP-148 Affects Membrane Integrity of E. hirae More Significantly Than OP-145
3.3. SAAP-148 Depolarizes E. hirae Cytoplasmic Membranes More Efficiently Than OP-145
3.4. SAAP-148 Permeabilizes E. hirae Cytoplasmic Membrane More Efficiently Than OP-145
3.5. SAAP-148 Interacts More Avidly with Membranes at the Molecular Level Than OP-145
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hancock, R.E.W.; Sahl, H.-G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Malanovic, N.; Lohner, K. Antimicrobial Peptides Targeting Gram-Positive Bacteria. Pharmaceuticals 2016, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Breij, A.; Riool, M.; Cordfunke, R.A.; Malanovic, N.; de Boer, L.; Koning, R.I.; Ravensbergen, E.; Franken, M.; van der Heijde, T.; Boekema, B.K.; et al. The antimicrobial peptide SAAP-148 combats drug-resistant bacteria and biofilms. Sci. Transl. Med. 2018, 10, eaan4044. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malanovic, N.; Marx, L.; Blondelle, S.E.; Pabst, G.; Semeraro, E.F. Experimental concepts for linking the biological activities of antimicrobial peptides to their molecular modes of action. Biochim. Biophys. Acta Biomembr. 2020, 1862, 183275. [Google Scholar] [CrossRef] [PubMed]
- Lohner, K. New strategies for novel antibiotics: Peptides targeting bacterial cell membranes. Gen. Physiol. Biophys. 2009, 28, 105–116. [Google Scholar] [CrossRef]
- Boman, H.G. Antibacterial peptides: Basic facts and emerging concepts. J. Intern. Med. 2003, 254, 197–215. [Google Scholar] [CrossRef]
- Wang, G.; Li, X.; Wang, Z. APD3: The antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44, D1087–D1093. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial Peptides: Diversity, Mechanism of Action and Strategies to Improve the Activity and Biocompatibility In Vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Dijksteel, G.S.; Ulrich, M.M.W.; Middelkoop, E.; Boekema, B.K.H.L. Review: Lessons Learned From Clinical Trials Using Antimicrobial Peptides (AMPs). Front. Microbiol. 2021, 12, 616979. [Google Scholar] [CrossRef]
- Ridyard, K.E.; Overhage, J. The Potential of Human Peptide LL-37 as an Antimicrobial and Anti-Biofilm Agent. Antibiotics 2021, 10, 650. [Google Scholar] [CrossRef]
- Haisma, E.M.; de Breij, A.; Chan, H.; van Dissel, J.T.; Drijfhout, J.W.; Hiemstra, P.S.; El Ghalbzouri, A.; Nibbering, P.H. LL-37-Derived Peptides Eradicate Multidrug-Resistant Staphylococcus aureus from Thermally Wounded Human Skin Equivalents. Antimicrob. Agents Chemother. 2014, 58, 4411–4419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nibbering, P.H.; Göblyös, A.; Adriaans, A.E.; Cordfunke, R.A.; Ravensbergen, B.; Rietveld, M.H.; Zwart, S.; Commandeur, S.; van Leeuwen, R.; Haisma, E.M.; et al. Eradication of meticillin-resistant Staphylococcus aureus from human skin by the novel LL-37-derived peptide P10 in four pharmaceutical ointments. Int. J. Antimicrob. Agents 2019, 54, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Nell, M.J.; Tjabringa, G.S.; Wafelman, A.R.; Verrijk, R.; Hiemstra, P.S.; Drijfhout, J.W.; Grote, J.J. Development of novel LL-37 derived antimicrobial peptides with LPS and LTA neutralizing and antimicrobial activities for therapeutic application. Peptides 2006, 27, 649–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, L.; Huang, J.-A. The Antibacterial Effects of Antimicrobial Peptides OP-145 against Clinically Isolated Multi-Resistant Strains. Jpn. J. Infect. Dis. 2017, 70, 601–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malanovic, N.; Leber, R.; Schmuck, M.; Kriechbaum, M.; Cordfunke, R.A.; Drijfhout, J.W.; de Breij, A.; Nibbering, P.H.; Kolb, D.; Lohner, K. Phospholipid-driven differences determine the action of the synthetic antimicrobial peptide OP-145 on Gram-positive bacterial and mammalian membrane model systems. Biochim. Biophyica Acta 2015, 1848, 2437–2447. [Google Scholar] [CrossRef] [Green Version]
- Peek, N.F.A.W.; Nell, M.J.; Brand, R.; Jansen-Werkhoven, T.; van Hoogdalem, E.J.; Verrijk, R.; Vonk, M.J.; Wafelman, A.R.; Valentijn, A.R.P.M.; Frijns, J.H.M.; et al. Ototopical drops containing a novel antibacterial synthetic peptide: Safety and efficacy in adults with chronic suppurative otitis media. PLoS ONE 2020, 15, e0231573. [Google Scholar] [CrossRef] [Green Version]
- Scheper, H.; Wubbolts, J.M.; Verhagen, J.A.M.; de Visser, A.W.; van der Wal, R.J.P.; Visser, L.G.; de Boer, M.G.J.; Nibbering, P.H. SAAP-148 Eradicates MRSA Persisters Within Mature Biofilm Models Simulating Prosthetic Joint Infection. Front. Microbiol. 2021, 12, 625952. [Google Scholar] [CrossRef]
- Oren, Z.; Lerman, J.C.; Gudmundsson, G.H.; Agerberth, B.; Shai, Y. Structure and organization of the human antimicrobial peptide LL-37 in phospholipid membranes: Relevance to the molecular basis for its non-cell-selective activity. Biochem. J. 1999, 341, 501–513. [Google Scholar] [CrossRef]
- Wildman, K.A.H.; Lee, D.-K.; Ramamoorthy, A. Mechanism of Lipid Bilayer Disruption by the Human Antimicrobial Peptide, LL-37. Biochemistry 2003, 42, 6545–6558. [Google Scholar] [CrossRef]
- Lohner, K. Membrane-active Antimicrobial Peptides as Template Structures for Novel Antibiotic Agents. Curr. Top. Med. Chem. 2017, 17, 508–519. [Google Scholar] [CrossRef]
- Malanovic, N.; Lohner, K. Gram-positive bacterial cell envelopes: The impact on the activity of antimicrobial peptides. Biochim. Biophys. Acta Biomembr. 2016, 1858, 936–946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevcsik, E.; Pabst, G.; Jilek, A.; Lohner, K. How lipids influence the mode of action of membrane-active peptides. Biochim. Biophys. Acta. 2007, 1768, 2586–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sevcsik, E.; Pabst, G.; Richter, W.; Danner, S.; Amenitsch, H.; Lohner, K. Interaction of LL-37 with Model Membrane Systems of Different Complexity: Influence of the Lipid Matrix. Biophys. J. 2008, 94, 4688–4699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sancho-Vaello, E.; Gil-Carton, D.; François, P.; Bonetti, E.-J.; Kreir, M.; Pothula, K.R.; Kleinekathöfer, U.; Zeth, K. The structure of the antimicrobial human cathelicidin LL-37 shows oligomerization and channel formation in the presence of membrane mimics. Sci. Rep. 2020, 10, 17356. [Google Scholar] [CrossRef]
- Scheenstra, M.R.; van den Belt, M.; Tjeerdsma-van Bokhoven, J.L.M.; Schneider, V.A.F.; Ordonez, S.R.; van Dijk, A.; Veldhuizen, E.J.A.; Haagsman, H.P. Cathelicidins PMAP-36, LL-37 and CATH-2 are similar peptides with different modes of action. Sci. Rep. 2019, 9, 4780. [Google Scholar] [CrossRef]
- Dorschner, R.A.; Lopez-Garcia, B.; Peschel, A.; Kraus, D.; Morikawa, K.; Nizet, V.; Gallo, R.L. The mammalian ionic environment dictates microbial susceptibility to antimicrobial defense peptides. FASEB J. 2006, 20, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Boaretti, M.; Canepari, P. Identification of daptomycin-binding proteins in the membrane of Enterococcus hirae. Antimicrob Agents Chemother 1995, 39, 2068–2072. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Maupetit, J.; Derreumaux, P.; Tufféry, P. Improved PEP-FOLD Approach for Peptide and Miniprotein Structure Prediction. J. Chem. Theory Comput. 2014, 10, 4745–4758. [Google Scholar] [CrossRef]
- Thevenet, P.; Shen, Y.; Maupetit, J.; Guyon, F.; Derreumaux, P.; Tuffery, P. PEP-FOLD: An updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012, 40, W288–W293. [Google Scholar] [CrossRef] [Green Version]
- Beaufays, J.; Lins, L.; Thomas, A.; Brasseur, R. In silico predictions of 3D structures of linear and cyclic peptides with natural and non-proteinogenic residues. J. Pept. Sci. 2012, 18, 17–24. [Google Scholar] [CrossRef]
- Snider, C.; Jayasinghe, S.; Hristova, K.; White, S.H. MPEx: A tool for exploring membrane proteins. Protein Sci. 2009, 18, 2624–2628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, S.H.; Wimley, W.C. MEMBRANE PROTEIN FOLDING AND STABILITY: Physical Principles. Annu. Rev. Biophys. Biomol. Struct. 1999, 28, 319–365. [Google Scholar] [CrossRef] [PubMed]
- Scheinpflug, K.; Krylova, O.; Nikolenko, H.; Thurm, C.; Dathe, M. Evidence for a Novel Mechanism of Antimicrobial Action of a Cyclic R-,W-Rich Hexapeptide. PLoS ONE 2015, 10, e0125056. [Google Scholar] [CrossRef]
- Wolinski, H.; Kohlwein, S.D. Microscopic and spectroscopic techniques to investigate lipid droplet formation and turnover in yeast. Methods Mol. Biol. 2015, 1270, 289–305. [Google Scholar] [CrossRef] [PubMed]
- Zweytick, D.; Japelj, B.; Mileykovskaya, E.; Zorko, M.; Dowhan, W.; Blondelle, S.E.; Riedl, S.; Jerala, R.; Lohner, K. N-acylated peptides derived from human lactoferricin perturb organization of cardiolipin and phosphatidylethanolamine in cell membranes and induce defects in Escherichia coli cell division. PLoS ONE 2014, 9, e90228. [Google Scholar] [CrossRef] [Green Version]
- Zweytick, D.; Deutsch, G.; Andrä, J.; Blondelle, S.E.; Vollmer, E.; Jerala, R.; Lohner, K. Studies on lactoferricin-derived Escherichia coli membrane-active peptides reveal differences in the mechanism of N-acylated versus nonacylated peptides. J. Biol. Chem. 2011, 286, 21266–21276. [Google Scholar] [CrossRef] [Green Version]
- Broekhuyse, R.M. Phospholipids in tissues of the eye. I. Isolation, characterization and quantitative analysis by two-dimensional thin-layer chromatography of diacyl and vinyl-ether phospholipids. Biochim. Biophys Acta 1968, 152, 307–315. [Google Scholar] [CrossRef]
- Freire, J.M.; Domingues, M.M.; Matos, J.; Melo, M.N.; Veiga, A.S.; Santos, N.C.; Castanho, M.A.R.B. Using zeta-potential measurements to quantify peptide partition to lipid membranes. Eur. Biophys. J. 2011, 40, 481–487. [Google Scholar] [CrossRef] [Green Version]
- Domingues, M.M.; Silva, P.M.; Franquelim, H.G.; Carvalho, F.A.; Castanho, M.A.R.B.; Santos, N.C. Antimicrobial protein rBPI21-induced surface changes on Gram-negative and Gram-positive bacteria. Nanomedicine 2014, 10, 543–551. [Google Scholar] [CrossRef]
- Malanovic, N.; Ön, A.; Pabst, G.; Zellner, A.; Lohner, K. Octenidine: Novel insights into the detailed killing mechanism of Gram-negative bacteria at a cellular and molecular level. Int. J. Antimicrob. Agents 2020, 56, 106146. [Google Scholar] [CrossRef]
- Koch, A.L. Bacterial Growth and Form; Springer Science + Business Media: Dordrecht, The Netherlands, 2013; ISBN 9789401708272. [Google Scholar]
- te Winkel, J.D.; Gray, D.A.; Seistrup, K.H.; Hamoen, L.W.; Strahl, H. Analysis of Antimicrobial-Triggered Membrane Depolarization Using Voltage Sensitive Dyes. Front. Cell Dev. Biol. 2016, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prenner, E.J.; Lewis, R.N.A.H.; Kondejewski, L.H.; Hodges, R.S.; McElhaney, R.N. Differential scanning calorimetric study of the effect of the antimicrobial peptide gramicidin S on the thermotropic phase behavior of phosphatidylcholine, phosphatidylethanolamine and phosphatidylglycerol lipid bilayer membranes. Biochim. Biophys. Acta (BBA) Biomembr. 1999, 1417, 211–223. [Google Scholar] [CrossRef] [Green Version]
- Benesch, M.G.; Lewis, R.N.; McElhaney, R.N. On the miscibility of cardiolipin with 1,2-diacyl phosphoglycerides: Binary mixtures of dimyristoylphosphatidylglycerol and tetramyristoylcardiolipin. Biochim. Biophys. Acta 2015, 1848, 2878–2888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lohner, K. DSC Studies on the Modulation of Membrane Lipid Polymorphism and Domain Organization by Antimicrobial Peptides. In Biocalorimetry: Foundations and Contemporary Approaches; Bastos, M., Ed.; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2015; pp. 169–190. ISBN 978-1-4822-4665-0. [Google Scholar]
- Fiore, E.; van Tyne, D.; Gilmore, M.S. Pathogenicity of Enterococci. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef] [PubMed]
- Anghinah, R.; Watanabe, R.G.S.; Simabukuro, M.M.; Guariglia, C.; Pinto, L.F.; Gonçalves, D.C.d.M.E. Native Valve Endocarditis due to Enterococcus hirae Presenting as a Neurological Deficit. Case Rep. Neurol. Med. 2013, 2013, 636070. [Google Scholar] [CrossRef]
- Dicpinigaitis, P.V.; de Aguirre, M.; Divito, J. Enterococcus hirae Bacteremia Associated with Acute Pancreatitis and Septic Shock. Case Rep. Infect. Dis. 2015, 2015, 123852. [Google Scholar] [CrossRef] [Green Version]
- Hancock, L.E.; Murray, B.E.; Sillanpää, J. Enterococcal Cell Wall Components and Structures. In Enterococci: From Commensals to Leading Causes of Drug Resistant Infection; Gilmore, M.S., Clewell, D.B., Ike, Y., Shankar, N., Eds.; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Shockman, G.D.; Martin, J.T. Autolytic enzyme system of Streptococcus faecalis. IV. Electron microscopic observations of autolysin and lysozyme action. J. Bacteriol. 1968, 96, 1803–1810. [Google Scholar] [CrossRef] [Green Version]
- Shockman, G.D. The autolytic (‘suicidase’) system of Enterococcus hirae: From lysine depletion autolysis to biochemical and molecular studies of the two muramidases of Enterococcus hirae ATCC 9790. Fems. Microbiol. Lett. 1992, 100, 261–267. [Google Scholar] [CrossRef]
- Liu, S.; Brul, S.; Zaat, S.A.J. Isolation of Persister Cells of Bacillus subtilis and Determination of Their Susceptibility to Antimicrobial Peptides. Int. J. Mol. Sci. 2021, 22, 59. [Google Scholar] [CrossRef]
- Yau, W.-M.; Wimley, W.C.; Gawrisch, A.K.; White, S.H. The Preference of Tryptophan for Membrane Interfaces. Biochemistry 1998, 37, 14713–14718. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; She, P.; Zhou, L.; Zeng, X.; Xu, L.; Liu, Y.; Chen, L.; Wu, Y. High-Throughput Identification of Antibacterials Against Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 591426. [Google Scholar] [CrossRef] [PubMed]







| Peptide | Concentration (µM) | Time (min) | |||||
|---|---|---|---|---|---|---|---|
| 5 | 10 | 20 | 30 | 60 | 120 | ||
| SAAP-148 | 0.1 | ||||||
| 0.2 | |||||||
| 0.4 | |||||||
| OP-145 | 0.8 | ||||||
| 1.6 | |||||||
| 3.2 | |||||||
| Tlow | ∆Hlow | Tpre | ∆Hpre | Tm | ∆Hm | |
|---|---|---|---|---|---|---|
| (°C) | (kcal/mol) | (°C) | (kcal/mol) | (°C) | (kcal/mol) | |
| PG | 12.0 | 1.0 | 22.9 | 7.9 | ||
| CL | 17.9 | 4.7 | 27.9 | 3.8 | 41.0 | 17.3 |
| PG/CL | 24.4 | 9.9 | ||||
| PG/CL + SAAP-148 | ||||||
| 1 mol% | 24.8 # | 10.1 * | ||||
| 4 mol% | 28.3 | 2.1 | ||||
| PG/CL + OP-145 | ||||||
| 1 mol% | 25.0 # | 10.1 * | ||||
| 4 mol% | 24.3; 29.1 | 16.1 * |
| E. hirae | E. hirae Membrane | Model Membranes | |||
|---|---|---|---|---|---|
| 99.9% killing | half-maximal depolarization | half-maximal permeability | maximal surface neutralization | maximal permeability | |
| SAAP-148 | 0.4 µM | 0.8 µM | 0.8 µM | >4 mol% | 0.5 mol% |
| OP-145 | 3.2 µM | 6.4 µM | >25.6 µM | >8 mol% | 8 mol% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piller, P.; Wolinski, H.; Cordfunke, R.A.; Drijfhout, J.W.; Keller, S.; Lohner, K.; Malanovic, N. Membrane Activity of LL-37 Derived Antimicrobial Peptides against Enterococcus hirae: Superiority of SAAP-148 over OP-145. Biomolecules 2022, 12, 523. https://doi.org/10.3390/biom12040523
Piller P, Wolinski H, Cordfunke RA, Drijfhout JW, Keller S, Lohner K, Malanovic N. Membrane Activity of LL-37 Derived Antimicrobial Peptides against Enterococcus hirae: Superiority of SAAP-148 over OP-145. Biomolecules. 2022; 12(4):523. https://doi.org/10.3390/biom12040523
Chicago/Turabian StylePiller, Paulina, Heimo Wolinski, Robert A. Cordfunke, Jan Wouter Drijfhout, Sandro Keller, Karl Lohner, and Nermina Malanovic. 2022. "Membrane Activity of LL-37 Derived Antimicrobial Peptides against Enterococcus hirae: Superiority of SAAP-148 over OP-145" Biomolecules 12, no. 4: 523. https://doi.org/10.3390/biom12040523
APA StylePiller, P., Wolinski, H., Cordfunke, R. A., Drijfhout, J. W., Keller, S., Lohner, K., & Malanovic, N. (2022). Membrane Activity of LL-37 Derived Antimicrobial Peptides against Enterococcus hirae: Superiority of SAAP-148 over OP-145. Biomolecules, 12(4), 523. https://doi.org/10.3390/biom12040523