The Ribosome Is the Ultimate Receptor for Trypsin Modulating Oostatic Factor (TMOF)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, and Chemicals
2.2. Cloning of Late Larval Ae. aegypti Trypsin
2.3. Inhibition of Luciferase Translation in S30 Bacterial Extract
2.4. Inhibition of Larval Ae. aegypti Late Trypsin Translation in Insect Cell Extract
2.5. Western Blotting and Mass Spectra Analyses
2.5.1. Western Blotting
2.5.2. Mass Spectra Analysis
2.6. Purification of E. coli Ribosomes
2.7. Binding of AeaTMOF-FITC to E. coli Ribosome Assay and Kinetics
2.8. Molecular Modeling and Docking
2.9. Effect of AeaTMOF and Oncocin112 (1–13) in Vivo on E. coli Growth
2.10. Statistical Analysis
3. Results
3.1. Inhibition of Luciferase Biosynthesis in E. coli S30 Extract System
3.1.1. Inhibition by AeaTMOF
3.1.2. Inhibition by Oncocin112 (1–13)
3.2. Inhibition of Ae. aegypti Late Larval Trypsin Biosynthesis with AeaTMOF
3.3. Binding Kinetics of AeaTMOF to E. coli Ribosomes
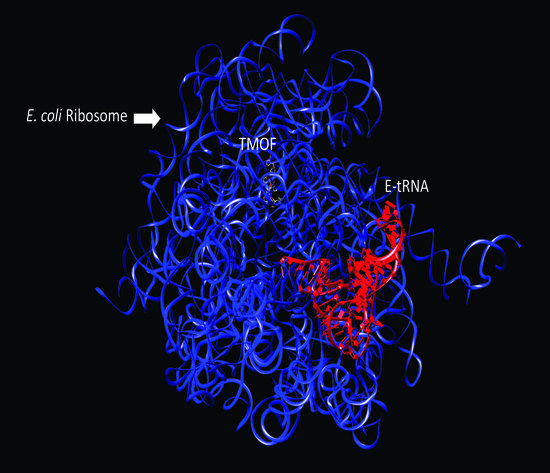
3.4. Three-Dimensional Modeling of AeaTMOF Binding to Bacterial Ribosome
3.4.1. Docking to T. thermophilus Ribosome
3.4.2. Docking to D. melanogaster and E. coli Ribosomes
3.5. Inhibition of E.coli Growth with AeaTMOF and Oncocin112 (1–13)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borovsky, D. Oostatic hormone inhibits biosynthesis of midgut proteolytic enzymes and egg development in mosquitoes. Arch. Insect Biochem. Physiol. 1988, 7, 187–210. [Google Scholar] [CrossRef]
- Borovsky, D.; Song, Q.; Ma, M.; Carlson, D.A. Biosynthesis, secretion and cytoimmunochemistry of trypsin modulating oostatic factor of Aedes aegypti. Arch. Insect Biochem. Physiol. 1994, 27, 27–38. [Google Scholar] [CrossRef]
- Borovsky, D.; Verhaert, P.; Rouge, P.; Powell, C.A.; De Loof, A. Blood digestion in Culex quinquefasciatus by trypsin is translationally regulated by Trypsin Modulating Oostatic Factor (TMOF). Front. Physiol. 2021, 12, 764061. [Google Scholar] [CrossRef]
- Borovsky, D.; Janssen, I.; Vanden Broeck, J.; Huybrechts, R.; Verhaert, P.; DeBondt, H.L.; Bylemans, D.; DeLoof, A. Molecular sequencing and modeling of Neobellieria bullata trypsin: Evidence for translational control with Neb TMOF. Eur. J. Biochem. 1996, 237, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, D.; Rabindran, S.; Dawson, W.O.; Powell, C.R.; Iannotti, D.; Morris, T.; Shabanowitz, J.; Hunt, D.F.; De Bondt, H.L.; De Loof, A. Expression of Aedes TMOF on the virion of TMV: Potential larvicide. Proc. Natl. Acad. Sci. USA 2006, 103, 18963–18968. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schoenberg, D.; Maquat, L.E. Regulation of cytoplasmic mRNA decay. Nat. Rev. Genet. 2012, 13, 246–259. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, D.; Deckers, K.; Vanhove, A.C.; Verstraete, M.; Rouge, P.; Shatters, R.G., Jr.; Powel, C.A. Cloning and Characterization of Aedes aegypti Trypsin Modulating Oostatic Factor (TMOF) Gut-Receptor. Biomolecules 2021, 11, 934. [Google Scholar] [CrossRef]
- Casteels, P.; Ampe, C.; Jacobs, F.; Vaeck, M.; Tempst, P. Apidaecins: Antibacterial peptides from honeybees. EMBO J. 1989, 8, 2387–2391. [Google Scholar] [CrossRef]
- Gennaro, R.; Zanetti, M.; Benincasa, M.; Podda, E.; Miani, M. Pro-rich antimicrobial peptides from animals:structure, biological functions and mechanism of action. Curr. Pharm. Des. 2002, 8, 763–778. [Google Scholar] [CrossRef]
- Li, J.X.; Xu, X.Q.; Yu, H.N.; Yang, H.L.; Huang, Z.X.; Lai, R. Direct antimicrobial activities of PR-bombesin. Life Sci. 2006, 78, 1953–1956. [Google Scholar] [CrossRef]
- Destoumieux, D.; Bulet, P.; Strub, J.M.; Van Dorsselaer, A.; Bachère, E. Recombinant expression and range of activity of penaeidins, antimicrobial peptides from penaeid shrimp. Eur. J. Biochem. 1999, 266, 335–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gueguen, Y.; Bernard, R.; Julie, F.; Paulina, S.; Delphine, D.G.; Franck, V.; Philippe, B.; Evelyne, B. Oyster hemocytes express a proline-rich peptide displaying synergistic antimicrobial activity with a defensin. Mol. Immunol. 2009, 46, 516–522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krizsan, A.; Volke, D.; Weinert, S.; Strater, N.; Knappe, D.; Hoffmann, R. Insect-derived proline-rich antimicrobial peptides kill bacteria by inhibiting bacterial protein translation at the 70 S ribosome. Angew. Chem. Int. Ed. Engl. 2014, 53, 12236–12239. [Google Scholar] [CrossRef] [PubMed]
- Seefeldt, A.C.; Nguyen, F.; Antunes, S.; Perebaskine, N.; Graf, M.; Arenz, S.; Inampudi, K.K.; Douat, C.; Guichard, G.; Wilson, D.N.; et al. The proline-rich antimicrobial peptide Onc112 inhibits translation by blocking and destabilizing the initiation complex. Nat. Struct. Mol. 2015, 22, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.N.; Lomakin, I.B.; Gagnon, M.G.; Steitz, T.A. The mechanism of inhibition of protein synthesis by the proline-rich peptide oncocin. Nat. Struct. Mol. Biol. 2015, 2, 466–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borovsky, D.; Carlson, D.A.; Griffin, P.R.; Shabanowitz, J.; Hunt, D.F. Mass Spectrometry and characterization of Aedes aegypti trypsin modulating oostatic factor (TMOF) and its analogs. Insect Biochem. Mol. Biol. 1993, 23, 703–712. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Van Ekert, E.; Heylen, K.; Rouge, P.; Powell, C.A.; Shatters, R.G., Jr.; Smagghe, G.; Borovsky, D. Aedes aegypti juvenile hormone acid methyltransferase, the ultimate enzyme in the biosynthetic pathway of juvenile hormone III, exhibits substrate control. J. Insect Physiol. 2014, 64, 62–73. [Google Scholar] [CrossRef]
- Mehta, P.; Woo, P.; Venkataraman, K.; Karzai, A.W. Ribosome purification approaches for studying interactions of regulatory proteins and RNAs with the ribosome. Methods Mol Biol. 2012, 905, 273–289. [Google Scholar] [CrossRef] [Green Version]
- Rivera, M.C.; Maguire, B.; Lake, J.A. Purification of 70S Ribosomes. Cold Spring Harb. Protoc. 2015, 2015, 300–302. [Google Scholar] [CrossRef]
- Curto, E.V.; Jarpe, M.A.; Blalock, J.B.; Borovsky, D.; Krishna, N.R. Solution structure of trypsin modulating oostatic factor is a left-handed helix. Biochem. Biophys. Res. Commun. 1993, 193, 688–693. [Google Scholar] [CrossRef]
- Krieger, E.; Koraimann, G.; Vriend, G. Increasing the precision of comparative models with YASARA NOVA—A self-parametrizing force field. Proteins 2002, 47, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Anger, A.M.; Armache, J.-P.; Berninghausen, O.; Habeck, M.; Subklewe, M.; Wilson, D.N.; Beckmazn, R. Structure of the human and Drosophila 80S ribosome. Nature 2013, 497, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Mattiuzzo, M.; Bandiera, A.; Gennaro, R.; Benincasa, M.; Pacor, S.; Antcheva, N.; Scocchi, M. Role of the Escherichia coli SbmA in the antimicrobialactivity of proline-rich peptides. Mol. Microbiol. 2007, 66, 151–163. [Google Scholar] [CrossRef]
- Borovsky, D. Trypsin modulating oostatic factor for developing resistant crops. In Insectidices Design Using Advanced Technologies; Ishaaya, I., Nauen, R., Horowitz, A.R., Eds.; Springer: Berlin, Germany, 2007. [Google Scholar]
- Borovsky, D.; Carlson, D.A.; Griffin, P.R.; Shabanowitz, J.; Hunt, D.F. Mosquito oostatic factor a novel decapeptide modulating trypsin-like enzyme biosynthesis in the midgut. FASEB J. 1990, 4, 3015–3020. [Google Scholar] [CrossRef]
- Borovsky, D.; Sterner, A.; Powell, C.A. Cloning and Expressing Trypsin Modulating Oostatic Factor (TMOF) in Chlorella desiccata to control Mosquito Larvae. Arch. Insect Biochem. Physiol. 2016, 91, 17–36. [Google Scholar] [CrossRef]
- Borovsky, D.; Nauwelaers, S.; Powell, C.A.; Shatters, R.G., Jr. Cloning, genetic engineering and characterization of TMOF expressed in Saccharomyces cerervisiae to control larval mosquitoes. J. Insect Physiol. 2018, 106, 134–146. [Google Scholar] [CrossRef]
- Borovsky, D.; Nauwelaers, S.; Shatters, R., Jr. Biochemical and molecular characterization of Pichia pastoris cells expressing multiple TMOF genes (tmfA) for mosquito larval control. Front. Physiol. 2020, 11, 527. [Google Scholar] [CrossRef]
- Ezure, T.; Suzuki, T.; Higashide, S.; Shintani, E.; Endo, K.; Kobayashi, S.-I.; Shikata, M.; Ito, M.; Tanimizu, K.; Nishimura, O. Cell-free protein synthesis system prepared from insect cells by freeze-thawing. Biotechnol. Prog. 2006, 22, 1570–1577. [Google Scholar] [CrossRef] [PubMed]
- Kearse, M.G.; Wilusz, J.E. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev. 2017, 31, 1717–1731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runti, G.; del Carmen Lopez Ruitz, M.; Stoilova, T.; Hussain, R.; Jennions, M.; Choudhury, H.G.; Benincasa, M.; Gennaro, R.; Beis, K.; Scocchi, M. Functional characterization of SbmA, a bacterial inner membrane transporter required for importing the antimicrobial peptide Bac7 (1–35). J. Bacteriol. 2013, 195, 5343–5351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Knappe, D.; Piantavigna, S.; Hansen, A.; Mechler, A.; Binas, A.; Nolte, O.; Martin, L.I. Oncocin (VDKPPYLPRPRPPPPIYNR-NH2): A novel antibacterial peptide optimized against gram-negative human pathogens. J. Med. Chem. 2010, 53, 5240–5247. [Google Scholar] [CrossRef] [PubMed]
- Borovsky, D.; Powell, C.A.; Nayar, J.K.; Blalock, J.E.; Hayes, T.K. Characterization and localization of mosquito-gut receptors for trypsin modulating oostatic factor (TMOF) using a complementary peptide and immunocytochemistry. FASEB J. 1994, 8, 350–355. [Google Scholar] [CrossRef]
- Zhu, Y.; Weisshaar, J.C.; Mustafi, M. Long-term effects of the proline-rich antimicrobial peptide Oncocin 112 on the Escherichia coli translation machinery. J. Biol. Chem. 2020, 295, 13314–13325. [Google Scholar] [CrossRef]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Borovsky, D.; Rougé, P.; Shatters, R.G., Jr. The Ribosome Is the Ultimate Receptor for Trypsin Modulating Oostatic Factor (TMOF). Biomolecules 2022, 12, 577. https://doi.org/10.3390/biom12040577
Borovsky D, Rougé P, Shatters RG Jr. The Ribosome Is the Ultimate Receptor for Trypsin Modulating Oostatic Factor (TMOF). Biomolecules. 2022; 12(4):577. https://doi.org/10.3390/biom12040577
Chicago/Turabian StyleBorovsky, Dov, Pierre Rougé, and Robert G. Shatters, Jr. 2022. "The Ribosome Is the Ultimate Receptor for Trypsin Modulating Oostatic Factor (TMOF)" Biomolecules 12, no. 4: 577. https://doi.org/10.3390/biom12040577
APA StyleBorovsky, D., Rougé, P., & Shatters, R. G., Jr. (2022). The Ribosome Is the Ultimate Receptor for Trypsin Modulating Oostatic Factor (TMOF). Biomolecules, 12(4), 577. https://doi.org/10.3390/biom12040577