Production of New Microbially Conjugated Bile Acids by Human Gut Microbiota
Abstract
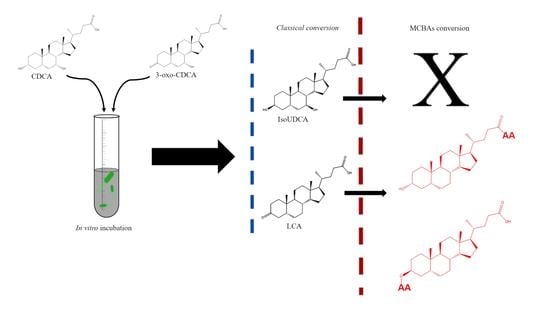
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Collection of Human Fecal Samples
2.3. Conversion Experiments by Human Fecal Cultures
2.4. UPLC-IM-QTOF-MS Analysis
2.5. Metabolomics Data Treatment
3. Results
3.1. Classical Microbial Conversion of Bile Acids
3.2. Production of Microbially Conjugated Bile Acids (MCBAs)
4. Discussion
5. Conclusions
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chiang, J.Y.L. Regulation of bile acid synthesis: Pathways, nuclear receptors, and mechanisms. J. Hepatol. 2004, 40, 539–551. [Google Scholar] [CrossRef] [PubMed]
- Monte, M.J.; Marin, J.J.G.; Antelo, A.; Vazquez-Tato, J. Bile acids: Chemistry, physiology, and pathophysiology. World J. Gastroenterol. WJG 2009, 15, 804–816. [Google Scholar] [CrossRef] [PubMed]
- Bjorkhem, I.; Araya, Z.; Rudling, M.; Angelin, B.; Einarsson, C.; Wikvall, K. Differences in the regulation of the classical and the alternative pathway for bile acid synthesis in human liver—No coordinate regulation of CYP7A1 and CYP27A1. J. Biol. Chem. 2002, 277, 26804–26807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallim, T.Q.D.; Tarling, E.J.; Edwards, P.A. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar] [CrossRef] [Green Version]
- Chiang, J.Y.L. Bile acid metabolism and signaling. Compr. Physiol. 2013, 3, 1191–1212. [Google Scholar] [CrossRef] [Green Version]
- Joyce, S.A.; Gahan, C.G.M. Bile acid modifications at the microbe-host interface: Potential for nutraceutical and pharmaceutical interventions in host health. Annu. Rev. Food Sci. Technol. 2016, 7, 313–333. [Google Scholar] [CrossRef]
- Gérard, P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013, 3, 14–24. [Google Scholar] [CrossRef] [Green Version]
- Long, S.L.; Gahan, C.G.M.; Joyce, S.A. Interactions between gut bacteria and bile in health and disease. Mol. Aspects Med. 2017, 56, 54–65. [Google Scholar] [CrossRef]
- Franco, P.; Porru, E.; Fiori, J.; Gioiello, A.; Cerra, B.; Roda, G.; Caliceti, C.; Simoni, P.; Roda, A. Identification and quantification of oxo-bile acids in human faeces with liquid chromatography-mass spectrometry: A potent tool for human gut acidic sterolbiome studies. J. Chromatogr. A 2019, 1585, 70–81. [Google Scholar] [CrossRef]
- Guzior, D.V.; Quinn, R.A. Review: Microbial transformations of human bile acids. Microbiome 2021, 9, 140. [Google Scholar] [CrossRef]
- Russell, D.W. The enzymes, regulation, and genetics of bile acid synthesis. Annu. Rev. Biochem. 2003, 72, 137–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oduyebo, I.; Camilleri, M. Bile acid disease: The emerging epidemic. Curr. Opin. Gastroen. 2017, 33, 189–195. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lajczak-McGinley, N.K.; Porru, E.; Fallon, C.M.; Smyth, J.; Curley, C.; McCarron, P.A.; Tambuwala, M.M.; Roda, A.; Keely, S.J. The secondary bile acids, ursodeoxycholic acid and lithocholic acid, protect against intestinal inflammation by inhibition of epithelial apoptosis. Physiol. Rep. 2020, 8, e14456. [Google Scholar] [CrossRef]
- Li, P.; Killinger, B.A.; Ensink, E.; Beddows, I.; Yilmaz, A.; Lubben, N.; Lamp, J.; Schilthuis, M.; Vega, I.E.; Woltjer, R.; et al. Gut microbiota dysbiosis is associated with elevated bile acids in Parkinson’s disease. Metabolites 2021, 11, 29. [Google Scholar] [CrossRef] [PubMed]
- García, C.J.; García-Villalba, R.; Moreno, D.A.; Tomás-Barberán, F.A. Gut microbiota interactions with dietary terpenoids and nitrogen-containing phytochemicals. Compr. Gut Microbiota 2022, 124–148. [Google Scholar] [CrossRef]
- Sagar, N.M.; Duboc, H.; Kay, G.L.; Alam, M.T.; Wicaksono, A.N.; Covington, J.A.; Quince, C.; Kokkorou, M.; Svolos, V.; Palmieri, L.J.; et al. The pathophysiology of bile acid diarrhoea: Differences in the colonic microbiome, metabolome and bile acids. Sci. Rep. 2020, 10, 20436. [Google Scholar] [CrossRef]
- Lloyd-Price, J.; Arze, C.; Ananthakrishnan, A.N.; Schirmer, M.; Avila-Pacheco, J.; Poon, T.W.; Andrews, E.; Ajami, N.J.; Bonham, K.S.; Brislawn, C.J.; et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019, 569, 655–662. [Google Scholar] [CrossRef]
- Distrutti, E.; Monaldi, L.; Ricci, P.; Fiorucci, S. Gut microbiota role in irritable bowel syndrome: New therapeutic strategies. World J. Gastroenterol. 2016, 22, 2219–2241. [Google Scholar] [CrossRef]
- Reiter, S.; Dunkel, A.; Metwaly, A.; Panes, J.; Salas, A.; Haller, D.; Hofmann, T. Development of a highly sensitive ultra-high-performance liquid chromatography coupled to electrospray ionization tandem mass spectrometry quantitation method for fecal Bile acids and application on Crohn’s disease studies. J. Agr. Food Chem. 2021, 69, 5238–5251. [Google Scholar] [CrossRef]
- Garcia, C.J.; Beltran, D.; Tomas-Barberan, F.A. Human gut microbiota metabolism of dietary sesquiterpene lactones: Untargeted metabolomics study of lactucopicrin and lactucin conversion in vitro and in vivo. Mol. Nutr. Food Res. 2020, 64, 2000619. [Google Scholar] [CrossRef]
- García-Villalba, R.; Beltrán, D.; Espín, J.C.; Selma, M.V.; Tomás-Barberán, F.A. Time course production of urolithins from ellagic acid by human gut microbiota. J. Agric. Food Chem. 2013, 61, 8797–8806. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones, B.V.; Begley, M.; Hill, C.; Gahan, C.G.; Marchesi, J.R. Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 13580–13585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joyce, S.A.; Gahan, C.G. Disease-associated changes in bile acid profiles and links to altered gut microbiota. Dig. Dis. 2017, 35, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Ridlon, J.M.; Hylemon, P.B. Identification and characterization of two bile acid coenzyme a transferases from Clostridium scindens, a bile acid 7α-dehydroxylating intestinal bacterium. J. Lipid Res. 2012, 53, 66–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirano, S.; Masuda, N. Epimerization of the 7-hydroxy group of bile acids by the combination of two kinds of microorganisms with 7 alpha- and 7 beta-hydroxysteroid dehydrogenase activity, respectively. J. Lipid Res. 1981, 22, 1060–1068. [Google Scholar] [CrossRef]
- Quinn, R.A.; Melnik, A.V.; Vrbanac, A.; Fu, T.; Patras, K.A.; Christy, M.P.; Bodai, Z.; Belda-Ferre, P.; Tripathi, A.; Chung, L.K.; et al. Global chemical effects of the microbiome include new bile-acid conjugations. Nature 2020, 579, 123–129. [Google Scholar] [CrossRef]
- Molinero, N.; Ruiz, L.; Sánchez, B.; Margolles, A.; Delgado, S. Intestinal bacteria interplay with bile and cholesterol metabolism: Implications on host physiology. Front. Physiol. 2019, 10, 185. [Google Scholar] [CrossRef] [Green Version]
- Hofmann, A.F. The enterohepatic circulation of bile acids in mammals: Form and functions. Front. Biosci. 2009, 14, 2584–2598. [Google Scholar] [CrossRef] [Green Version]
- Perez, M.J.; Briz, O. Bile-acid-induced cell injury and protection. World J. Gastroenterol. WJG 2009, 15, 1677–1689. [Google Scholar] [CrossRef]
- Kelsey, M.I.; Molina, J.E.; Shingkwan, S.H.; Kinkai, H. The identification of microbial metabolites of sulfolithocholic acid. J. Lipid Res. 1980, 21, 751–759. [Google Scholar] [CrossRef]
- Benson, G.M.; Haskins, N.J.; Eckers, C.; Moore, P.J.; Reid, D.G.; Mitchell, R.C.; Waghmare, S.; Suckling, K.E. Polydeoxycholate in human and hamster feces—A major product of cholate metabolism. J. Lipid Res. 1993, 34, 2121–2134. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Harris, S.C.; Bhowmik, S.; Kang, D.J.; Hylemon, P.B. Consequences of bile salt biotransformations by intestinal bacteria. Gut Microbes 2016, 7, 22–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, M.J.; Carey, M.C. The hydrophobic-hydrophilic balance of bile salts. Inverse correlation between reverse-phase high performance liquid chromatographic mobilities and micellar cholesterol-solubilizing capacities. J. Lipid Res. 1982, 23, 70–80. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile salt biotransformations by human intestinal bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biot. 2017, 101, 47–64. [Google Scholar] [CrossRef] [Green Version]
- Thomas, C.; Pellicciari, R.; Pruzanski, M.; Auwerx, J.; Schoonjans, K. Targeting bile-acid signalling for metabolic diseases. Nat. Rev. Drug Discov. 2008, 7, 678–693. [Google Scholar] [CrossRef]
- Huang, F.; Zheng, X.; Ma, X.; Jiang, R.; Zhou, W.; Zhou, S.; Zhang, Y.; Lei, S.; Wang, S.; Kuang, J.; et al. Theabrownin from Pu-erh tea attenuates hypercholesterolemia via modulation of gut microbiota bile acid metabolism. Nat. Commun. 2009, 10, 17. [Google Scholar] [CrossRef] [Green Version]
- Van Faassen, A.; Tangerman, A.; Bueno-de-Mesquita, B.H. Serum bile acids and risk factors for colorectal cancer. Br. J. Cancer 2004, 90, 632–634. [Google Scholar] [CrossRef] [Green Version]
- Chow, M.D.; Lee, Y.H.; Guo, G.L. The role of bile acids in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Mol. Aspects Med. 2017, 56, 34–44. [Google Scholar] [CrossRef]
- Kessoku, T.; Kobayashi, T.; Imajo, K.; Honda, Y.; Ogawa, Y.; Higurashi, T.; Kato, S.; Yoneda, M.; Oikawa, Y.; Tanaka, Y.; et al. Increased levels of bile acid in feces plays an important role in pathophysiology of non-alcoholic steatohepatitis. J. Hepatol. 2019, 70, e534. [Google Scholar] [CrossRef]
- Ferrell, J.M.; Chiang, J.Y.L. Understanding bile acid signaling in diabetes: From pathophysiology to therapeutic targets. Diabetes Metab. 2019, 43, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Zhan, K.; Zheng, H.; Li, J.; Wu, H.; Qin, S.; Luo, L.; Huang, S. Gut Microbiota-bile acid crosstalk in diarrhea-irritable bowel syndrome. Biomed. Res. Int. 2020, 2020, 20203828249. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Wolf, P.G.; Gaskins, H.R. Taurocholic acid metabolism by gut microbes and colon cancer. Gut Microbes 2016, 7, 201–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Yang, Z.; Shi, Z.; Zhu, Z.; Li, C.; Du, Z.; Zhang, Y.; Wang, Z.; Jiao, Z.; Tian, X.; et al. Analysis of bile acid profile in plasma to differentiate cholangiocarcinoma from benign biliary diseases and healthy controls. J. Steroid Biochem. Mol. Biol. 2021, 205, 105775. [Google Scholar] [CrossRef] [PubMed]
- Perino, A.; Demagny, H.; Velazquez-Villegas, L.A.; Schoonjans, K. Molecular Physiology of Bile Acid Signaling in Health, Disease, and Aging. Physiol. Rev. 2021, 101, 683–731. [Google Scholar] [CrossRef] [PubMed]





| Compound Name | Formula | m/z | Rt | MS/MS Fragments | Collision E | Abundance abs * |
|---|---|---|---|---|---|---|
| Chenodeoxycholic acid (CDCA) | C40H40O4 | 391.2849 | 14.33 | 391.2864; 373.2752 | 40 | 1.9 × 105 |
| Isoursochenodexycholic acid (iUDCA) | C40H40O4 | 391.2849 | 11.66 | 391.2855; 373.2784 | 40 | 2.6 × 107 |
| 3-oxo-chenodeoxycholic acid (3-oxoCDCA) | C40H38O4 | 389.2692 | 14.58 | 389.2708; 345.2819; 343.2666; 371.2643 | 40 | 2.1 × 105 |
| 7-oxo-lithocholic acid (7-oxoLCA) | C40H38O4 | 389.2692 | 13.29 | 389.2722; 345.2814; 343.2668 | 40 | 3.1 × 104 |
| Lithocholic acid (LCA) | C40H40O3 | 375.2899 | 18.03 | 375.2944; 357.2838; 355.2679 | 50 | 9.5 × 107 |
| 7α-Hydroxy-5β-cholan-24-oic acid | C40H40O3 | 375.2899 | 17.15 | 375.2947; 357.2947 | 50 | 1.4 × 107 |
| 3-oxo-5β-cholan-24-oic-acid | C40H38O3 | 373.2743 | 18.4 | 373.2732; 355.2626 | 30 | 1.7 × 107 |
| Compound Name | m/z | Formula | Rt | MS/MS Fragment |
|---|---|---|---|---|
| Leucolithocholic; Leucolithocholate ester | 488.3754 | C30H51NO4 | 18.50 | 488.3739; 130.0872 |
| Leucolithocholic; Leucolithocholate ester | 488.3735 | C30H51NO4 | 18.17 | 488.3813; 444.3902; 130.0867 |
| Isoleucolithocholic; Isoleucolithocholate ester | 488.3745 | C30H51NO4 | 17.20 | 488.3808; 444.3317; 130.0875 |
| Valolithocholic; Valoisolithocholic; Valoisolithocholate ester | 474.3602 | C29H49NO4 | 17.15 | 474.3640; 430.3761; 116.0709 |
| Valolithocholic; Valoisolithocholic; Valoisolithocholate ester | 474.3591 | C29H49NO4 | 16.80 | 474.3650; 430.3731; 116.0714 |
| Valolithocholic; Valoisolithocholic; Valoisolithocholate ester | 474.3548 | C29H49NO4 | 16.16 | 474.3638; 430.3730; 116.0713 |
| Valolithocholic; Valoisolithocholic; Valoisolithocholate ester | 474.3589 | C29H49NO4 | 15.48 | 474.3643; 430.3727; 116.0716 |
| Valolithocholic; Valoisolithocholic; Valoisolithocholate ester | 474.3584 | C29H49NO4 | 14.83 | 474.3652; 430.3729; 116.0712 |
| Triptophano-dioxochenodeoxycholic | 573.3340 | C35H46N2O5 | 15.60 | N/D |
| Leucochenodeoxycholic; isolecochenodeoxycholic; Leucolithocholate ester | 504.3694 | C30H51NO5 | 13.90 | N/D |
| Prololithocholic; Prololithocholate ester; valo-oxolithocholic | 472.3412 | C29H47NO4 | 18.20 | N/D |
| Alanolithocholic; Alanolithocholate ester; Serocholic acid; Serocholate ester | 446.3285 | C27H45NO4 | 15.58 | N/D |
| Alanolithocholic; Alanolithocholate ester; Serocholic acid; Serocholate ester | 446.3269 | C27H45NO4 | 15.90 | N/D |
| Alanolithocholic; Alanolithocholate ester; Serocholic acid; Serocholate ester | 446.3261 | C27H45NO4 | 15.92 | N/D |
| Arginolithocholic; Arginolithocholate ester | 531.3759 | C30H52N4O4 | 15.50 | N/D |
| Lysocholic acid; Lysocholate ester | 487.3930 | C30H52N2O3 | 14.89 | N/D |
| Lysocholic acid; Lysocholate ester | 487.3933 | C30H52N2O3 | 14.28 | N/D |
| Threonocholic acid; Threonocholate ester | 460.3432 | C28H47NO4 | 15.15 | N/D |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia, C.J.; Kosek, V.; Beltrán, D.; Tomás-Barberán, F.A.; Hajslova, J. Production of New Microbially Conjugated Bile Acids by Human Gut Microbiota. Biomolecules 2022, 12, 687. https://doi.org/10.3390/biom12050687
Garcia CJ, Kosek V, Beltrán D, Tomás-Barberán FA, Hajslova J. Production of New Microbially Conjugated Bile Acids by Human Gut Microbiota. Biomolecules. 2022; 12(5):687. https://doi.org/10.3390/biom12050687
Chicago/Turabian StyleGarcia, Carlos J., Vit Kosek, David Beltrán, Francisco A. Tomás-Barberán, and Jana Hajslova. 2022. "Production of New Microbially Conjugated Bile Acids by Human Gut Microbiota" Biomolecules 12, no. 5: 687. https://doi.org/10.3390/biom12050687
APA StyleGarcia, C. J., Kosek, V., Beltrán, D., Tomás-Barberán, F. A., & Hajslova, J. (2022). Production of New Microbially Conjugated Bile Acids by Human Gut Microbiota. Biomolecules, 12(5), 687. https://doi.org/10.3390/biom12050687