Vitamin A Deficiency Alters the Phototransduction Machinery and Distinct Non-Vision-Specific Pathways in the Drosophila Eye Proteome
Abstract
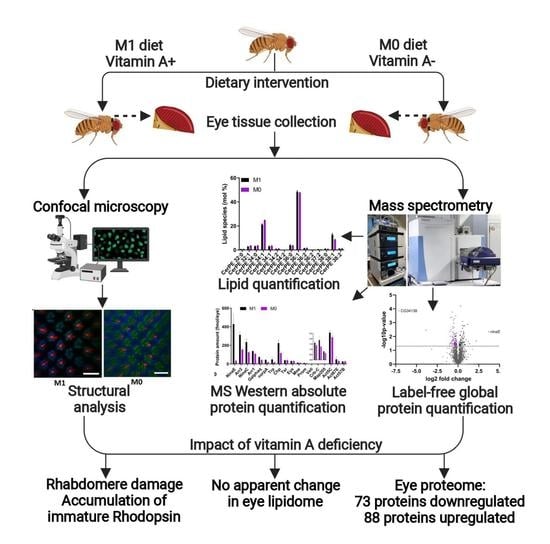
:1. Introduction
2. Materials and Methods
2.1. Food Media
2.2. Drosophila Melanogaster Stocks
2.3. Imaging the Drosophila Eye
2.4. Confocal Microscopy and Immunohistochemistry of Drosophila Photoreceptors
2.5. Rhabdomere Measurements and Statistics
2.6. Lipid Extraction and Shotgun Lipidomics on the Drosophila Eye
2.7. MS Western and Global Proteome Analysis of the Drosophila Eye by GeLC-MS/MS
2.8. Data Processing for Protein Identification and Quantification
2.9. Functional Annotation and Enrichment Analysis
3. Results
3.1. Wild Type Eye and Rhabdomere Morphology of Drosophila Raised on a Minimal Synthetic Diet
3.2. Vitamin A Deficiency Affects Rhabdomere Volume and Rhodopsin Expression
3.3. Vitamin A Deficiency Has No Apparent Effects on the Lipidome of the Drosophila Eye
3.4. Vitamin A Deficiency Alters the Phototransduction Machinery
3.5. Vitamin A Deficiency Alters Non-Vision-Specific Pathways of the Drosophila Eye Proteome
4. Discussion
4.1. Vitamin A Deficiency Decreases the Expression of Most Phototransduction Proteins
4.2. Vitamin A Deficiency Affects Non-Vision-Specific Pathways of the Eye Proteome
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sommer, A. Vitamin a deficiency and clinical disease: An historical overview. J. Nutr. 2008, 138, 1835–1839. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewett, D.; Labaf, M.; Lam-Kamath, K.; Zarringhalam, K.; Rister, J. Vitamin A deficiency affects gene expression in the Drosophila melanogaster head. G3 2021, 11, jkab297. [Google Scholar] [CrossRef] [PubMed]
- Saari, J.C. Vitamin A and Vision. Subcell. Biochem. 2016, 81, 231–259. [Google Scholar] [CrossRef] [PubMed]
- von Lintig, J. Metabolism of carotenoids and retinoids related to vision. J. Biol. Chem. 2012, 287, 1627–1634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nichols, R.; Pak, W.L. Characterization of Drosophila melanogaster rhodopsin. J. Biol. Chem. 1985, 260, 12670–12674. [Google Scholar] [CrossRef]
- Harris, W.A.; Ready, D.F.; Lipson, E.D.; Hudspeth, A.J.; Stark, W.S. Vitamin A deprivation and Drosophila photopigments. Nature 1977, 266, 648–650. [Google Scholar] [CrossRef] [PubMed]
- Randall, A.S.; Liu, C.H.; Chu, B.; Zhang, Q.; Dongre, S.A.; Juusola, M.; Franze, K.; Wakelam, M.J.; Hardie, R.C. Speed and sensitivity of phototransduction in Drosophila depend on degree of saturation of membrane phospholipids. J. Neurosci. 2015, 35, 2731–2746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiefer, C.; Sumser, E.; Wernet, M.F.; Von Lintig, J. A class B scavenger receptor mediates the cellular uptake of carotenoids in Drosophila. Proc. Natl. Acad. Sci. USA 2002, 99, 10581–10586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oberhauser, V.; Voolstra, O.; Bangert, A.; von Lintig, J.; Vogt, K. NinaB combines carotenoid oxygenase and retinoid isomerase activity in a single polypeptide. Proc. Natl. Acad. Sci. USA 2008, 105, 19000–19005. [Google Scholar] [CrossRef] [Green Version]
- Vogt, K.; Kirschfeld, K. Chemical identity of the chromophores of fly visual pigment. Science 1984, 71, 211–213. [Google Scholar] [CrossRef]
- von Lintig, J.; Vogt, K. Filling the gap in vitamin A research. Molecular identification of an enzyme cleaving beta-carotene to retinal. J. Biol. Chem. 2000, 275, 11915–11920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Isono, K.; Tanimura, T.; Oda, Y.; Tsukahara, Y. Dependency on light and vitamin A derivatives of the biogenesis of 3-hydroxyretinal and visual pigment in the compound eyes of Drosophila melanogaster. J. Gen. Physiol. 1988, 92, 587–600. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, K.; Nagatani, H.; Ozaki, M.; Tokunaga, F. Maturation of major Drosophila rhodopsin, ninaE, requires chromophore 3-hydroxyretinal. Neuron 1993, 10, 1113–1119. [Google Scholar] [CrossRef]
- Huber, A.; Wolfrum, U.; Paulsen, R. Opsin maturation and targeting to rhabdomeral photoreceptor membranes requires the retinal chromophore. Eur. J. Cell Biol. 1994, 63, 219–229. [Google Scholar] [PubMed]
- Cunningham, T.J.; Duester, G. Mechanisms of retinoic acid signalling and its roles in organ and limb development. Nat. Rev. Mol. Cell Biol. 2015, 16, 110–123. [Google Scholar] [CrossRef] [Green Version]
- Oro, A.E.; McKeown, M.; Evans, R.M. Relationship between the product of the Drosophila ultraspiracle locus and the vertebrate retinoid X receptor. Nature 1990, 347, 298–301. [Google Scholar] [CrossRef]
- Bonneton, F.; Zelus, D.; Iwema, T.; Robinson-Rechavi, M.; Laudet, V. Rapid divergence of the ecdysone receptor in Diptera and Lepidoptera suggests coevolution between ECR and USP-RXR. Mol. Biol. Evol. 2003, 20, 541–553. [Google Scholar] [CrossRef] [Green Version]
- Kam, R.K.; Deng, Y.; Chen, Y.; Zhao, H. Retinoic acid synthesis and functions in early embryonic development. Cell Biosci. 2012, 2, 11. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.D.; Thomas, C.F.; Marietta, R.G.; Stark, W.S. Vitamin A, visual pigments, and visual receptors in Drosophila. Microsc Res. Tech. 1996, 35, 418–430. [Google Scholar] [CrossRef]
- Schwudke, D.; Liebisch, G.; Herzog, R.; Schmitz, G.; Shevchenko, A. Shotgun lipidomics by tandem mass spectrometry under data-dependent acquisition control. Meth. Enzymol. 2007, 433, 175–191. [Google Scholar] [CrossRef]
- Schwudke, D.; Schuhmann, K.; Herzog, R.; Bornstein, S.R.; Shevchenko, A. Shotgun lipidomics on high resolution mass spectrometers. Cold Spring Harb Perspect. Biol. 2011, 3, a004614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vasilj, A.; Gentzel, M.; Ueberham, E.; Gebhardt, R.; Shevchenko, A. Tissue proteomics by one-dimensional gel electrophoresis combined with label-free protein quantification. J. Proteome Res. 2012, 11, 3680–3689. [Google Scholar] [CrossRef] [PubMed]
- Kumar, M.; Joseph, S.R.; Augsburg, M.; Bogdanova, A.; Drechsel, D.; Vastenhouw, N.L.; Buchholz, F.; Gentzel, M.; Shevchenko, A. MS Western, a method of multiplexed absolute protein quantification is a practical alternative to Western blotting. Mol. Cell Proteomics 2018, 17, 384–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsiao, H.Y.; Johnston, R.J.; Jukam, D.; Vasiliauskas, D.; Desplan, C.; Rister, J. Dissection and immunohistochemistry of larval, pupal and adult Drosophila retinas. J. Vis. Exp. 2012, e4347. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, M.; Sampaio, J.L.; Palm, W.; Brankatschk, M.; Eaton, S.; Shevchenko, A. Effects of diet and development on the Drosophila lipidome. Mol. Syst. Biol. 2012, 8, 600. [Google Scholar] [CrossRef] [PubMed]
- Hebbar, S.; Schuhmann, K.; Shevchenko, A.; Knust, E. Hydroxylated sphingolipid biosynthesis regulates photoreceptor apical domain morphogenesis. J. Cell Biol. 2020, 219, e201911100. [Google Scholar] [CrossRef]
- Matyash, V.; Liebisch, G.; Kurzchalia, T.V.; Shevchenko, A.; Schwudke, D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008, 49, 1137–1146. [Google Scholar] [CrossRef] [Green Version]
- Liebisch, G.; Binder, M.; Schifferer, R.; Langmann, T.; Schulz, B.; Schmitz, G. High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim. Biophys. Acta. 2006, 1761, 121–128. [Google Scholar] [CrossRef]
- Herzog, R.; Schwudke, D.; Schuhmann, K.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011, 12, R8. [Google Scholar] [CrossRef] [Green Version]
- Herzog, R.; Schuhmann, K.; Schwudke, D.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. LipidXplorer: A software for consensual cross-platform lipidomics. PLoS ONE 2012, 7, e29851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghuraman, B.K.; Hebbar, S.; Kumar, M.; Moon, H.; Henry, I.; Knust, E.; Shevchenko, A. Absolute Quantification of Proteins in the Eye of Drosophila melanogaster. Proteomics 2020, 20, e1900049. [Google Scholar] [CrossRef] [PubMed]
- Shevchenko, A.; Tomas, H.; Havlis, J.; Olsen, J.V.; Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 2006, 1, 2856–2860. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Knittelfelder, O.; Prince, E.; Sales, S.; Fritzsche, E.; Wohner, T.; Brankatschk, M.; Shevchenko, A. Sterols as dietary markers for Drosophila melanogaster. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2020, 1865, 158683. [Google Scholar] [CrossRef]
- Luersen, K.; Roder, T.; Rimbach, G. Drosophila melanogaster in nutrition research-the importance of standardizing experimental diets. Genes Nutr. 2019, 14, 3. [Google Scholar] [CrossRef]
- Rister, J.; Desplan, C. The retinal mosaics of opsin expression in invertebrates and vertebrates. Dev. Neurobiol. 2011, 71, 1212–1226. [Google Scholar] [CrossRef] [Green Version]
- Yau, K.W.; Hardie, R.C. Phototransduction motifs and variations. Cell 2009, 139, 246–264. [Google Scholar] [CrossRef] [Green Version]
- Chevesich, J.; Kreuz, A.J.; Montell, C. Requirement for the PDZ domain protein, INAD, for localization of the TRP store-operated channel to a signaling complex. Neuron 1997, 18, 95–105. [Google Scholar] [CrossRef] [Green Version]
- Schneuwly, S.; Burg, M.G.; Lending, C.; Perdew, M.H.; Pak, W.L. Properties of photoreceptor-specific phospholipase C encoded by the norpA gene of Drosophila melanogaster. J. Biol. Chem. 1991, 266, 24314–24319. [Google Scholar] [CrossRef]
- Hardie, R.C.; Peretz, A.; Suss-Toby, E.; Rom-Glas, A.; Bishop, S.A.; Selinger, Z.; Minke, B. Protein kinase C is required for light adaptation in Drosophila photoreceptors. Nature 1993, 363, 634–637. [Google Scholar] [CrossRef] [PubMed]
- Porter, J.A.; Yu, M.; Doberstein, S.K.; Pollard, T.D.; Montell, C. Dependence of calmodulin localization in the retina on the NINAC unconventional myosin. Science 1993, 262, 1038–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mecklenburg, K.L.; Takemori, N.; Komori, N.; Chu, B.; Hardie, R.C.; Matsumoto, H.; O’Tousa, J.E. Retinophilin is a light-regulated phosphoprotein required to suppress photoreceptor dark noise in Drosophila. J. Neurosci. 2010, 30, 1238–1249. [Google Scholar] [CrossRef] [Green Version]
- Venkatachalam, K.; Wasserman, D.; Wang, X.; Li, R.; Mills, E.; Elsaesser, R.; Li, H.S.; Montell, C. Dependence on a retinophilin/myosin complex for stability of PKC and INAD and termination of phototransduction. J. Neurosci. 2010, 30, 11337–11345. [Google Scholar] [CrossRef]
- Dolph, P.J.; Ranganathan, R.; Colley, N.J.; Hardy, R.W.; Socolich, M.; Zuker, C.S. Arrestin function in inactivation of G protein-coupled receptor rhodopsin in vivo. Science 1993, 260, 1910–1916. [Google Scholar] [CrossRef]
- Mazzotta, G.; Rossi, A.; Leonardi, E.; Mason, M.; Bertolucci, C.; Caccin, L.; Spolaore, B.; Martin, A.J.; Schlichting, M.; Grebler, R.; et al. Fly cryptochrome and the visual system. Proc. Natl. Acad. Sci. USA 2013, 110, 6163–6168. [Google Scholar] [CrossRef] [Green Version]
- Stuart, A.E. From fruit flies to barnacles, histamine is the neurotransmitter of arthropod photoreceptors. Neuron 1999, 22, 431–433. [Google Scholar] [CrossRef] [Green Version]
- Melzig, J.; Burg, M.; Gruhn, M.; Pak, W.L.; Buchner, E. Selective histamine uptake rescues photo- and mechanoreceptor function of histidine decarboxylase-deficient Drosophila mutant. J. Neurosci. 1998, 18, 7160–7166. [Google Scholar] [CrossRef] [Green Version]
- Satoh, A.K.; Ready, D.F. Arrestin1 mediates light-dependent rhodopsin endocytosis and cell survival. Curr. Biol. 2005, 15, 1722–1733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurudev, N.; Yuan, M.; Knust, E. chaoptin, prominin, eyes shut and crumbs form a genetic network controlling the apical compartment of Drosophila photoreceptor cells. BIO 2014, 3, 332–341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reinke, R.; Krantz, D.E.; Yen, D.; Zipursky, S.L. Chaoptin, a cell surface glycoprotein required for Drosophila photoreceptor cell morphogenesis, contains a repeat motif found in yeast and human. Cell 1988, 52, 291–301. [Google Scholar] [CrossRef]
- Zelhof, A.C.; Hardy, R.W.; Becker, A.; Zuker, C.S. Transforming the architecture of compound eyes. Nature 2006, 443, 696–699. [Google Scholar] [CrossRef] [PubMed]
- Tepass, U.; Theres, C.; Knust, E. crumbs encodes an EGF-like protein expressed on apical membranes of Drosophila epithelial cells and required for organization of epithelia. Cell 1990, 61, 787–799. [Google Scholar] [CrossRef]
- Rajala, R.V.S. Signaling roles of phosphoinositides in the retina. J. Lipid Res. 2021, 62, 100041. [Google Scholar] [CrossRef]
- Hardie, R.C.; Raghu, P.; Moore, S.; Juusola, M.; Baines, R.A.; Sweeney, S.T. Calcium influx via TRP channels is required to maintain PIP2 levels in Drosophila photoreceptors. Neuron 2001, 30, 149–159. [Google Scholar] [CrossRef]
- Smith, D.P.; Ranganathan, R.; Hardy, R.W.; Marx, J.; Tsuchida, T.; Zuker, C.S. Photoreceptor deactivation and retinal degeneration mediated by a photoreceptor-specific protein kinase C. Science 1991, 254, 1478–1484. [Google Scholar] [CrossRef]
- Shieh, B.H.; Zhu, M.Y.; Lee, J.K.; Kelly, I.M.; Bahiraei, F. Association of INAD with NORPA is essential for controlled activation and deactivation of Drosophila phototransduction in vivo. Proc. Natl. Acad. Sci. USA 1997, 94, 12682–12687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakrishnan, S.S.; Basu, U.; Shinde, D.; Thakur, R.; Jaiswal, M.; Raghu, P. Regulation of PI4P levels by PI4KIIIalpha during G-protein-coupled PLC signaling in Drosophila photoreceptors. J. Cell Sci. 2018, 131, jcs217257. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.H.; Bollepalli, M.K.; Long, S.V.; Asteriti, S.; Tan, J.; Brill, J.A.; Hardie, R.C. Genetic dissection of the phosphoinositide cycle in Drosophila photoreceptors. J. Cell Sci. 2018, 131, jcs214478. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, E.A.; Amato, C.; Fortier, T.M.; Velentzas, P.; Wood, W.; Baehrecke, E.H. A conserved myotubularin-related phosphatase regulates autophagy by maintaining autophagic flux. J. Cell Biol. 2020, 219, e201909073. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Reyes, I.; Chandel, N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat. Commun. 2020, 11, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cinege, G.; Zsamboki, J.; Vidal-Quadras, M.; Uv, A.; Csordas, G.; Honti, V.; Gabor, E.; Hegedus, Z.; Varga, G.I.B.; Kovacs, A.L.; et al. Genes encoding cuticular proteins are components of the Nimrod gene cluster in Drosophila. Insect Biochem. Mol. Biol. 2017, 87, 45–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Komori, N.; Usukura, J.; Matsumoto, H. Drosocrystallin, a major 52 kDa glycoprotein of the Drosophila melanogaster corneal lens. Purification, biochemical characterization, and subcellular localization. J. Cell Sci. 1992, 102 Pt 2, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Stahl, A.L.; Baucom, R.S.; Cook, T.A.; Buschbeck, E.K. A Complex Lens for a Complex Eye. Integr. Comp. Biol. 2017, 57, 1071–1081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dewett, D.; Lam-Kamath, K.; Poupault, C.; Khurana, H.; Rister, J. Mechanisms of vitamin A metabolism and deficiency in the mammalian and fly visual system. Dev. Biol. 2021, 476, 68–78. [Google Scholar] [CrossRef]
- Hammerling, U. Vitamin A as PKC Co-factor and Regulator of Mitochondrial Energetics. Subcell. Biochem. 2016, 81, 201–230. [Google Scholar] [CrossRef]
- Huang, H.W.; Brown, B.; Chung, J.; Domingos, P.M.; Ryoo, H.D. highroad Is a Carboxypetidase Induced by Retinoids to Clear Mutant Rhodopsin-1 in Drosophila Retinitis Pigmentosa Models. Cell Rep. 2018, 22, 1384–1391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.W.; Ryoo, H.D. Drosophila fabp is required for light-dependent Rhodopsin-1 clearance and photoreceptor survival. PLoS Genet. 2021, 17, e1009551. [Google Scholar] [CrossRef]
- Colley, N.J.; Cassill, J.A.; Baker, E.K.; Zuker, C.S. Defective intracellular transport is the molecular basis of rhodopsin-dependent dominant retinal degeneration. Proc. Natl. Acad. Sci. USA 1995, 92, 3070–3074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kurada, P.; O’Tousa, J.E. Retinal degeneration caused by dominant rhodopsin mutations in Drosophila. Neuron 1995, 14, 571–579. [Google Scholar] [CrossRef] [Green Version]
- Ryoo, H.D.; Domingos, P.M.; Kang, M.J.; Steller, H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007, 26, 242–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, M.J.; Ryoo, H.D. Suppression of retinal degeneration in Drosophila by stimulation of ER-associated degradation. Proc. Natl. Acad. Sci. USA 2009, 106, 17043–17048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryoo, H.D. Drosophila as a model for unfolded protein response research. BMB Rep. 2015, 48, 445–453. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pellikka, M.; Tanentzapf, G.; Pinto, M.; Smith, C.; McGlade, C.J.; Ready, D.F.; Tepass, U. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 2002, 416, 143–149. [Google Scholar] [CrossRef]
- Harding, H.P.; Zhang, Y.; Zeng, H.; Novoa, I.; Lu, P.D.; Calfon, M.; Sadri, N.; Yun, C.; Popko, B.; Paules, R.; et al. An integrated stress response regulates amino acid metabolism and resistance to oxidative stress. Mol. Cell 2003, 11, 619–633. [Google Scholar] [CrossRef]
- Han, J.; Back, S.H.; Hur, J.; Lin, Y.H.; Gildersleeve, R.; Shan, J.; Yuan, C.L.; Krokowski, D.; Wang, S.; Hatzoglou, M.; et al. ER-stress-induced transcriptional regulation increases protein synthesis leading to cell death. Nat. Cell Biol. 2013, 15, 481–490. [Google Scholar] [CrossRef]
- Rosenbaum, E.E.; Hardie, R.C.; Colley, N.J. Calnexin is essential for rhodopsin maturation, Ca2+ regulation, and photoreceptor cell survival. Neuron 2006, 49, 229–241. [Google Scholar] [CrossRef] [Green Version]
- Insinna, C.; Daniele, L.L.; Davis, J.A.; Larsen, D.D.; Kuemmel, C.; Wang, J.; Nikonov, S.S.; Knox, B.E.; Pugh, E.N., Jr. An S-opsin knock-in mouse (F81Y) reveals a role for the native ligand 11-cis-retinal in cone opsin biosynthesis. J. Neurosci. 2012, 32, 8094–8104. [Google Scholar] [CrossRef]
- Lobanova, E.S.; Finkelstein, S.; Skiba, N.P.; Arshavsky, V.Y. Proteasome overload is a common stress factor in multiple forms of inherited retinal degeneration. Proc. Natl. Acad. Sci. USA 2013, 110, 9986–9991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leonard, D.S.; Bowman, V.D.; Ready, D.F.; Pak, W.L. Degeneration of photoreceptors in rhodopsin mutants of Drosophila. J. Neurobio. 1992, 23, 605–626. [Google Scholar] [CrossRef] [PubMed]
- Jodeiri Farshbaf, M.; Kiani-Esfahani, A. Succinate dehydrogenase: Prospect for neurodegenerative diseases. Mitochondrion 2018, 42, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Van Vranken, J.G.; Bricker, D.K.; Dephoure, N.; Gygi, S.P.; Cox, J.E.; Thummel, C.S.; Rutter, J. SDHAF4 promotes mitochondrial succinate dehydrogenase activity and prevents neurodegeneration. Cell Metab. 2014, 20, 241–252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno, C.; Santos, R.M.; Burns, R.; Zhang, W.C. Succinate Dehydrogenase and Ribonucleic Acid Networks in Cancer and Other Diseases. Cancers 2020, 12, 3237. [Google Scholar] [CrossRef]
- Rowe, A.A.; Patel, P.D.; Gordillo, R.; Wert, K.J. Replenishment of TCA cycle intermediates provides photoreceptor resilience against neurodegeneration during progression of retinitis pigmentosa. JCI Insight 2021, 6, e150898. [Google Scholar] [CrossRef]
- Acin-Perez, R.; Hoyos, B.; Zhao, F.; Vinogradov, V.; Fischman, D.A.; Harris, R.A.; Leitges, M.; Wongsiriroj, N.; Blaner, W.S.; Manfredi, G.; et al. Control of oxidative phosphorylation by vitamin A illuminates a fundamental role in mitochondrial energy homoeostasis. FASEB J. 2010, 24, 627–636. [Google Scholar] [CrossRef] [Green Version]
- Hammerling, U. Retinol as electron carrier in redox signaling, a new frontier in vitamin A research. Hepatobiliary Surg. Nutr. 2016, 5, 15–28. [Google Scholar] [CrossRef]
- Gong, J.; Hoyos, B.; Acin-Perez, R.; Vinogradov, V.; Shabrova, E.; Zhao, F.; Leitges, M.; Fischman, D.; Manfredi, G.; Hammerling, U. Two protein kinase C isoforms, delta and epsilon, regulate energy homeostasis in mitochondria by transmitting opposing signals to the pyruvate dehydrogenase complex. FASEB J. 2012, 26, 3537–3549. [Google Scholar] [CrossRef] [Green Version]
- Hoyos, B.; Acin-Perez, R.; Fischman, D.A.; Manfredi, G.; Hammerling, U. Hiding in plain sight: Uncovering a new function of vitamin A in redox signaling. Biochim. Biophys. Acta. 2012, 1821, 241–247. [Google Scholar] [CrossRef]
- Chiu, H.J.; Fischman, D.A.; Hammerling, U. Vitamin A depletion causes oxidative stress, mitochondrial dysfunction, and PARP-1-dependent energy deprivation. FASEB J. 2008, 22, 3878–3887. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gilbert, C. The eye signs of vitamin A deficiency. Community Eye Health 2013, 26, 66–67. [Google Scholar] [PubMed]







| Pathways | Components |
|---|---|
| Phototransduction | NinaE/Rh1, Galphaq, Arr1, Arr2, InaC, InaD, NorpA, Trp |
| Phosphatidylinositol signaling | NorpA, PI4KIIIalpha, Ttc7, InaC, Ipp, Mtmr6 |
| Citrate/TCA cycle | ScsbetaA, SdhA, Kdn, ATPsynbeta, Men-b |
| Genetic information processing | Hsp22, Hsp23, Hsc70-3, Cnx99A, Gp93, CCT5, Wbl |
| Transport and catabolism | Npc1a, Gga, Ppt1, Rb |
| Metabolism of proteins | LeuRS, LysRS, MetRS, ThrRS, HisRS, IleRS, ValRS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kumar, M.; Has, C.; Lam-Kamath, K.; Ayciriex, S.; Dewett, D.; Bashir, M.; Poupault, C.; Schuhmann, K.; Knittelfelder, O.; Raghuraman, B.K.; et al. Vitamin A Deficiency Alters the Phototransduction Machinery and Distinct Non-Vision-Specific Pathways in the Drosophila Eye Proteome. Biomolecules 2022, 12, 1083. https://doi.org/10.3390/biom12081083
Kumar M, Has C, Lam-Kamath K, Ayciriex S, Dewett D, Bashir M, Poupault C, Schuhmann K, Knittelfelder O, Raghuraman BK, et al. Vitamin A Deficiency Alters the Phototransduction Machinery and Distinct Non-Vision-Specific Pathways in the Drosophila Eye Proteome. Biomolecules. 2022; 12(8):1083. https://doi.org/10.3390/biom12081083
Chicago/Turabian StyleKumar, Mukesh, Canan Has, Khanh Lam-Kamath, Sophie Ayciriex, Deepshe Dewett, Mhamed Bashir, Clara Poupault, Kai Schuhmann, Oskar Knittelfelder, Bharath Kumar Raghuraman, and et al. 2022. "Vitamin A Deficiency Alters the Phototransduction Machinery and Distinct Non-Vision-Specific Pathways in the Drosophila Eye Proteome" Biomolecules 12, no. 8: 1083. https://doi.org/10.3390/biom12081083
APA StyleKumar, M., Has, C., Lam-Kamath, K., Ayciriex, S., Dewett, D., Bashir, M., Poupault, C., Schuhmann, K., Knittelfelder, O., Raghuraman, B. K., Ahrends, R., Rister, J., & Shevchenko, A. (2022). Vitamin A Deficiency Alters the Phototransduction Machinery and Distinct Non-Vision-Specific Pathways in the Drosophila Eye Proteome. Biomolecules, 12(8), 1083. https://doi.org/10.3390/biom12081083