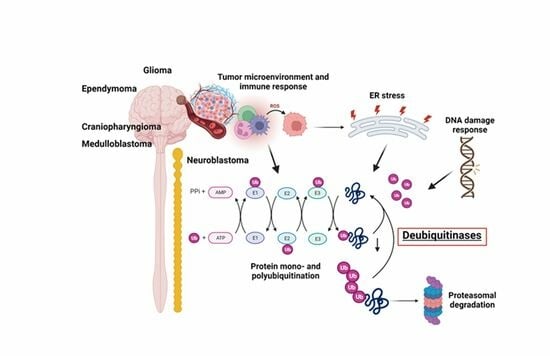
DUBing Primary Tumors of the Central Nervous System: Regulatory Roles of Deubiquitinases
Abstract
:1. Introduction
2. Methods
3. Results
3.1. Adult Glioma Show Differential Expression of DUBs
3.2. Chromosome 10 and DUB Expression in Astrocytic glioma
3.3. Differential Expression of DUBs in GBM Subtypes
3.4. Relationship between DUB Expression and Survival (French Dataset)
3.5. Role of DUBs in ER Stress and ERAD Signaling in Glioma
3.6. DUBs in the Regulation of Immune Responses in Glioma
3.7. DUBs and DNA Repair in Glioma
3.8. DUBs in Ependymoma
3.9. DUBs in Craniopharyngioma
3.10. DUB Regulation of Immune Response in Craniopharyngioma
3.11. DUBs and Medulloblastoma
3.12. Survival in Medulloblastoma Subgroups and DUB Expression
3.13. Medulloblastoma DUBs and ERAD
3.14. Medulloblastoma DUBs and the Immune Response
3.15. Medulloblastoma DUBs and DNA Damage Repair
3.16. DUBs in Neuroblastoma (Fischer Dataset)
4. Discussion
5. Summary and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, T.; Liu, Z.; Wang, Y.; Cheng, H.; Yang, Q.; Guo, A.; Ren, J.; Xue, Y. UUCD: A family-based database of ubiquitin and ubiquitin-like conjugation. Nucleic Acids Res. 2013, 41, D445–D451. [Google Scholar] [CrossRef]
- van der Veen, A.G.; Ploegh, H.L. Ubiquitin-like proteins. Annu. Rev. Biochem. 2012, 81, 323–357. [Google Scholar] [CrossRef]
- Varshavsky, A. The Ubiquitin System, Autophagy, and Regulated Protein Degradation. Annu. Rev. Biochem. 2017, 86, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I.; Schulman, B.A. An expanded lexicon for the ubiquitin code. Nat. Rev. Mol. Cell Biol. 2023, 24, 273–287. [Google Scholar] [CrossRef] [PubMed]
- Swatek, K.N.; Komander, D. Ubiquitin modifications. Cell Res. 2016, 26, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Tracz, M.; Bialek, W. Beyond K48 and K63: Non-canonical protein ubiquitination. Cell. Mol. Biol. Lett. 2021, 26, 1. [Google Scholar] [CrossRef]
- Jacobson, A.D.; Zhang, N.Y.; Xu, P.; Han, K.J.; Noone, S.; Peng, J.; Liu, C.W. The lysine 48 and lysine 63 ubiquitin conjugates are processed differently by the 26 s proteasome. J. Biol. Chem. 2009, 284, 35485–35494. [Google Scholar] [CrossRef]
- Kwon, Y.T.; Ciechanover, A. The Ubiquitin Code in the Ubiquitin-Proteasome System and Autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef]
- Dosa, A.; Csizmadia, T. The role of K63-linked polyubiquitin in several types of autophagy. Biol. Futur. 2022, 73, 137–148. [Google Scholar] [CrossRef]
- Liu, Z.; Gong, Z.; Jiang, W.X.; Yang, J.; Zhu, W.K.; Guo, D.C.; Zhang, W.P.; Liu, M.L.; Tang, C. Lys63-linked ubiquitin chain adopts multiple conformational states for specific target recognition. eLife 2015, 4, e05767. [Google Scholar] [CrossRef]
- Suresh, H.G.; Pascoe, N.; Andrews, B. The structure and function of deubiquitinases: Lessons from budding yeast. Open Biol. 2020, 10, 200279. [Google Scholar] [CrossRef] [PubMed]
- Mevissen, T.E.T.; Komander, D. Mechanisms of Deubiquitinase Specificity and Regulation. Annu. Rev. Biochem. 2017, 86, 159–192. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Clague, M.J.; Urbe, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 2009, 10, 550–563. [Google Scholar] [CrossRef]
- Lange, S.M.; Armstrong, L.A.; Kulathu, Y. Deubiquitinases: From mechanisms to their inhibition by small molecules. Mol. Cell 2022, 82, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Meng, T.; Chen, L.; Wei, W.; Wang, P. The role of ubiquitination in tumorigenesis and targeted drug discovery. Signal Transduct. Target. Ther. 2020, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Davis, M.I.; Simeonov, A. Ubiquitin-Specific Proteases as Druggable Targets. Drug Target Rev. 2015, 2, 60–64. [Google Scholar]
- Rong, C.; Zhou, R.; Wan, S.; Su, D.; Wang, S.L.; Hess, J. Ubiquitin Carboxyl-Terminal Hydrolases and Human Malignancies: The Novel Prognostic and Therapeutic Implications for Head and Neck Cancer. Front. Oncol. 2020, 10, 592501. [Google Scholar] [CrossRef]
- Johnston, S.C.; Riddle, S.M.; Cohen, R.E.; Hill, C.P. Structural basis for the specificity of ubiquitin C-terminal hydrolases. EMBO J. 1999, 18, 3877–3887. [Google Scholar] [CrossRef]
- Larsen, C.N.; Price, J.S.; Wilkinson, K.D. Substrate binding and catalysis by ubiquitin C-terminal hydrolases: Identification of two active site residues. Biochemistry 1996, 35, 6735–6744. [Google Scholar] [CrossRef]
- Makarova, K.S.; Aravind, L.; Koonin, E.V. A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem. Sci. 2000, 25, 50–52. [Google Scholar] [CrossRef]
- Du, J.; Fu, L.; Sui, Y.; Zhang, L. The function and regulation of OTU deubiquitinases. Front. Med. 2020, 14, 542–563. [Google Scholar] [CrossRef]
- Schluter, D.; Schulze-Niemand, E.; Stein, M.; Naumann, M. Ovarian tumor domain proteases in pathogen infection. Trends Microbiol. 2022, 30, 22–33. [Google Scholar] [CrossRef] [PubMed]
- Zeng, C.; Zhao, C.; Ge, F.; Li, Y.; Cao, J.; Ying, M.; Lu, J.; He, Q.; Yang, B.; Dai, X.; et al. Machado-Joseph Deubiquitinases: From Cellular Functions to Potential Therapy Targets. Front. Pharmacol. 2020, 11, 1311. [Google Scholar] [CrossRef] [PubMed]
- Patterson-Fortin, J.; Shao, G.; Bretscher, H.; Messick, T.E.; Greenberg, R.A. Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J. Biol. Chem. 2010, 285, 30971–30981. [Google Scholar] [CrossRef]
- Pan, X.; Wu, S.; Wei, W.; Chen, Z.; Wu, Y.; Gong, K. Structural and Functional Basis of JAMM Deubiquitinating Enzymes in Disease. Biomolecules 2022, 12, 910. [Google Scholar] [CrossRef] [PubMed]
- Dubiel, W.; Chaithongyot, S.; Dubiel, D.; Naumann, M. The COP9 Signalosome: A Multi-DUB Complex. Biomolecules 2020, 10, 1082. [Google Scholar] [CrossRef] [PubMed]
- Abdul Rehman, S.A.; Kristariyanto, Y.A.; Choi, S.Y.; Nkosi, P.J.; Weidlich, S.; Labib, K.; Hofmann, K.; Kulathu, Y. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol. Cell 2016, 63, 146–155. [Google Scholar] [CrossRef]
- Hickey, C.M.; Wilson, N.R.; Hochstrasser, M. Function and regulation of SUMO proteases. Nat. Rev. Mol. Cell Biol. 2012, 13, 755–766. [Google Scholar] [CrossRef]
- Tokarz, P.; Wozniak, K. SENP Proteases as Potential Targets for Cancer Therapy. Cancers 2021, 13, 2059. [Google Scholar] [CrossRef]
- Mendoza, H.M.; Shen, L.N.; Botting, C.; Lewis, A.; Chen, J.; Ink, B.; Hay, R.T. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 2003, 278, 25637–25643. [Google Scholar] [CrossRef]
- Qu, J.; Zou, T.; Lin, Z. The Roles of the Ubiquitin-Proteasome System in the Endoplasmic Reticulum Stress Pathway. Int. J. Mol. Sci. 2021, 22, 1526. [Google Scholar] [CrossRef]
- Shibata, Y.; Voeltz, G.K.; Rapoport, T.A. Rough sheets and smooth tubules. Cell 2006, 126, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.S.; Blower, M.D. The endoplasmic reticulum: Structure, function and response to cellular signaling. Cell. Mol. Life Sci. 2016, 73, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Almanza, A.; Carlesso, A.; Chintha, C.; Creedican, S.; Doultsinos, D.; Leuzzi, B.; Luis, A.; McCarthy, N.; Montibeller, L.; More, S.; et al. Endoplasmic reticulum stress signalling—From basic mechanisms to clinical applications. FEBS J. 2019, 286, 241–278. [Google Scholar] [CrossRef] [PubMed]
- He, B. Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Differ. 2006, 13, 393–403. [Google Scholar] [CrossRef]
- Shen, J.; Chen, X.; Hendershot, L.; Prywes, R. ER stress regulation of ATF6 localization by dissociation of BiP/GRP78 binding and unmasking of Golgi localization signals. Dev. Cell 2002, 3, 99–111. [Google Scholar] [CrossRef]
- Bertolotti, A.; Zhang, Y.; Hendershot, L.M.; Harding, H.P.; Ron, D. Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol. 2000, 2, 326–332. [Google Scholar] [CrossRef]
- Madden, E.; Logue, S.E.; Healy, S.J.; Manie, S.; Samali, A. The role of the unfolded protein response in cancer progression: From oncogenesis to chemoresistance. Biol. Cell 2019, 111, 1–17. [Google Scholar] [CrossRef]
- Park, J.; Cho, J.; Song, E.J. Ubiquitin-proteasome system (UPS) as a target for anticancer treatment. Arch. Pharmacal Res. 2020, 43, 1144–1161. [Google Scholar] [CrossRef]
- Jurkovicova, D.; Neophytou, C.M.; Gasparovic, A.C.; Goncalves, A.C. DNA Damage Response in Cancer Therapy and Resistance: Challenges and Opportunities. Int. J. Mol. Sci. 2022, 23, 14672. [Google Scholar] [CrossRef] [PubMed]
- Pilger, D.; Seymour, L.W.; Jackson, S.P. Interfaces between cellular responses to DNA damage and cancer immunotherapy. Genes Dev. 2021, 35, 602–618. [Google Scholar] [CrossRef]
- Vriend, J.; Klonisch, T. Genes of the Ubiquitin Proteasome System Qualify as Differential Markers in Malignant Glioma of Astrocytic and Oligodendroglial Origin. Cell. Mol. Neurobiol. 2023, 43, 1425–1452. [Google Scholar] [CrossRef] [PubMed]
- Vriend, J.; Thanasupawat, T.; Sinha, N.; Klonisch, T. Ubiquitin Proteasome Gene Signatures in Ependymoma Molecular Subtypes. Int. J. Mol. Sci. 2022, 23, 12330. [Google Scholar] [CrossRef] [PubMed]
- Maris, J.M.; Hogarty, M.D.; Bagatell, R.; Cohn, S.L. Neuroblastoma. Lancet 2007, 369, 2106–2120. [Google Scholar] [CrossRef] [PubMed]
- Park, J.R.; Bagatell, R.; London, W.B.; Maris, J.M.; Cohn, S.L.; Mattay, K.K.; Hogarty, M.; on behalf of the COG Neuroblastoma Committee. Children’s Oncology Group’s 2013 blueprint for research: Neuroblastoma. Pediatr. Blood Cancer 2013, 60, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ramakrishna, S.; Kim, K.S. Critical Roles of Deubiquitinating Enzymes in the Nervous System and Neurodegenerative Disorders. Mol. Cells 2020, 43, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Maksoud, S. The Role of the Ubiquitin Proteasome System in Glioma: Analysis Emphasizing the Main Molecular Players and Therapeutic Strategies Identified in Glioblastoma Multiforme. Mol. Neurobiol. 2021, 58, 3252–3269. [Google Scholar] [CrossRef]
- Sonoda, Y.; Ozawa, T.; Aldape, K.D.; Deen, D.F.; Berger, M.S.; Pieper, R.O. Akt pathway activation converts anaplastic astrocytoma to glioblastoma multiforme in a human astrocyte model of glioma. Cancer Res. 2001, 61, 6674–6678. [Google Scholar]
- Molinaro, A.M.; Taylor, J.W.; Wiencke, J.K.; Wrensch, M.R. Genetic and molecular epidemiology of adult diffuse glioma. Nat. Rev. Neurol. 2019, 15, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Park, J.W.; Lee, J.H. Genetic Architectures and Cell-of-Origin in Glioblastoma. Front. Oncol. 2020, 10, 615400. [Google Scholar] [CrossRef] [PubMed]
- Ruda, R.; Bruno, F.; Pellerino, A.; Soffietti, R. Ependymoma: Evaluation and Management Updates. Curr. Oncol. Rep. 2022, 24, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Bou Zerdan, M.; Assi, H.I. Oligodendroglioma: A Review of Management and Pathways. Front. Mol. Neurosci. 2021, 14, 722396. [Google Scholar] [CrossRef]
- Williamson, D.; Schwalbe, E.C.; Hicks, D.; Aldinger, K.A.; Lindsey, J.C.; Crosier, S.; Richardson, S.; Goddard, J.; Hill, R.M.; Castle, J.; et al. Medulloblastoma group 3 and 4 tumors comprise a clinically and biologically significant expression continuum reflecting human cerebellar development. Cell Rep. 2022, 40, 111162. [Google Scholar] [CrossRef] [PubMed]
- Funakoshi, Y.; Sugihara, Y.; Uneda, A.; Nakashima, T.; Suzuki, H. Recent advances in the molecular understanding of medulloblastoma. Cancer Sci. 2023, 114, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Rechberger, J.S.; Toll, S.A.; Vanbilloen, W.J.F.; Daniels, D.J.; Khatua, S. Exploring the Molecular Complexity of Medulloblastoma: Implications for Diagnosis and Treatment. Diagnostics 2023, 13, 2398. [Google Scholar] [CrossRef]
- Alomari, A.K.; Kelley, B.J.; Damisah, E.; Marks, A.; Hui, P.; DiLuna, M.; Vortmeyer, A. Craniopharyngioma arising in a Rathke’s cleft cyst: Case report. J. Neurosurg. Pediatr. 2015, 15, 250–254. [Google Scholar] [CrossRef]
- Johnsen, J.I.; Dyberg, C.; Wickstrom, M. Neuroblastoma—A Neural Crest Derived Embryonal Malignancy. Front. Mol. Neurosci. 2019, 12, 9. [Google Scholar] [CrossRef]
- Qiu, B.; Matthay, K.K. Advancing therapy for neuroblastoma. Nat. Rev. Clin. Oncol. 2022, 19, 515–533. [Google Scholar] [CrossRef]
- Mallepalli, S.; Gupta, M.K.; Vadde, R. Neuroblastoma: An Updated Review on Biology and Treatment. Curr. Drug Metab. 2019, 20, 1014–1022. [Google Scholar] [CrossRef]
- Chung, C.; Boterberg, T.; Lucas, J.; Panoff, J.; Valteau-Couanet, D.; Hero, B.; Bagatell, R.; Hill-Kayser, C.E. Neuroblastoma. Pediatr. Blood Cancer 2021, 68 (Suppl. S2), e28473. [Google Scholar] [CrossRef]
- Gravendeel, L.A.; Kouwenhoven, M.C.; Gevaert, O.; de Rooi, J.J.; Stubbs, A.P.; Duijm, J.E.; Daemen, A.; Bleeker, F.E.; Bralten, L.B.; Kloosterhof, N.K.; et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009, 69, 9065–9072. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef] [PubMed]
- Weishaupt, H.; Johansson, P.; Sundstrom, A.; Lubovac-Pilav, Z.; Olsson, B.; Nelander, S.; Swartling, F.J. Batch-normalization of cerebellar and medulloblastoma gene expression datasets utilizing empirically defined negative control genes. Bioinformatics 2019, 35, 3357–3364. [Google Scholar] [CrossRef] [PubMed]
- Ackermann, S.; Cartolano, M.; Hero, B.; Welte, A.; Kahlert, Y.; Roderwieser, A.; Bartenhagen, C.; Walter, E.; Gecht, J.; Kerschke, L.; et al. A mechanistic classification of clinical phenotypes in neuroblastoma. Science 2018, 362, 1165–1170. [Google Scholar] [CrossRef]
- Seifert, M.; Schackert, G.; Temme, A.; Schrock, E.; Deutsch, A.; Klink, B. Molecular Characterization of Astrocytoma Progression Towards Secondary Glioblastomas Utilizing Patient-Matched Tumor Pairs. Cancers 2020, 12, 1696. [Google Scholar] [CrossRef]
- Zhang, P.; Xiao, Z.; Wang, S.; Zhang, M.; Wei, Y.; Hang, Q.; Kim, J.; Yao, F.; Rodriguez-Aguayo, C.; Ton, B.N.; et al. ZRANB1 Is an EZH2 Deubiquitinase and a Potential Therapeutic Target in Breast Cancer. Cell Rep. 2018, 23, 823–837. [Google Scholar] [CrossRef]
- Duan, R.; Du, W.; Guo, W. EZH2: A novel target for cancer treatment. J. Hematol. Oncol. 2020, 13, 104. [Google Scholar] [CrossRef]
- Straining, R.; Eighmy, W. Tazemetostat: EZH2 Inhibitor. J. Adv. Pract. Oncol. 2022, 13, 158–163. [Google Scholar] [CrossRef]
- Fujisawa, H.; Reis, R.M.; Nakamura, M.; Colella, S.; Yonekawa, Y.; Kleihues, P.; Ohgaki, H. Loss of heterozygosity on chromosome 10 is more extensive in primary (de novo) than in secondary glioblastomas. Lab. Investig. 2000, 80, 65–72. [Google Scholar] [CrossRef]
- Balesaria, S.; Brock, C.; Bower, M.; Clark, J.; Nicholson, S.K.; Lewis, P.; de Sanctis, S.; Evans, H.; Peterson, D.; Mendoza, N.; et al. Loss of chromosome 10 is an independent prognostic factor in high-grade gliomas. Br. J. Cancer 1999, 81, 1371–1377. [Google Scholar] [CrossRef]
- Wiles, B.; Miao, M.; Coyne, E.; Larose, L.; Cybulsky, A.V.; Wing, S.S. USP19 deubiquitinating enzyme inhibits muscle cell differentiation by suppressing unfolded-protein response signaling. Mol. Biol. Cell 2015, 26, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Kato, M.; Nakamura, N. USP19-Mediated Deubiquitination Facilitates the Stabilization of HRD1 Ubiquitin Ligase. Int. J. Mol. Sci. 2016, 17, 1829. [Google Scholar] [CrossRef]
- Nagai, A.; Kadowaki, H.; Maruyama, T.; Takeda, K.; Nishitoh, H.; Ichijo, H. USP14 inhibits ER-associated degradation via interaction with IRE1α. Biochem. Biophys. Res. Commun. 2009, 379, 995–1000. [Google Scholar] [CrossRef] [PubMed]
- Blount, J.R.; Burr, A.A.; Denuc, A.; Marfany, G.; Todi, S.V. Ubiquitin-specific protease 25 functions in Endoplasmic Reticulum-associated degradation. PLoS ONE 2012, 7, e36542. [Google Scholar] [CrossRef]
- Zhu, D.; Xu, R.; Huang, X.; Tang, Z.; Tian, Y.; Zhang, J.; Zheng, X. Deubiquitinating enzyme OTUB1 promotes cancer cell immunosuppression via preventing ER-associated degradation of immune checkpoint protein PD-L1. Cell Death Differ. 2021, 28, 1773–1789. [Google Scholar] [CrossRef]
- Wu, Q.; Huang, Y.; Gu, L.; Chang, Z.; Li, G.M. OTUB1 stabilizes mismatch repair protein MSH2 by blocking ubiquitination. J. Biol. Chem. 2021, 296, 100466. [Google Scholar] [CrossRef] [PubMed]
- Unda, B.K.; Chalil, L.; Yoon, S.; Kilpatrick, S.; Irwin, C.; Xing, S.; Murtaza, N.; Cheng, A.; Brown, C.; Afonso, A.; et al. Impaired OTUD7A-dependent Ankyrin regulation mediates neuronal dysfunction in mouse and human models of the 15q13.3 microdeletion syndrome. Mol. Psychiatry 2023, 28, 1747–1769. [Google Scholar] [CrossRef]
- Yin, J.; Chen, W.; Chao, E.S.; Soriano, S.; Wang, L.; Wang, W.; Cummock, S.E.; Tao, H.; Pang, K.; Liu, Z.; et al. Otud7a Knockout Mice Recapitulate Many Neurological Features of 15q13.3 Microdeletion Syndrome. Am. J. Hum. Genet. 2018, 102, 296–308. [Google Scholar] [CrossRef]
- Bonacci, T.; Emanuele, M.J. Dissenting degradation: Deubiquitinases in cell cycle and cancer. Semin. Cancer Biol. 2020, 67, 145–158. [Google Scholar] [CrossRef]
- Das, T.; Chen, Z.; Hendriks, R.W.; Kool, M. A20/Tumor Necrosis Factor α-Induced Protein 3 in Immune Cells Controls Development of Autoinflammation and Autoimmunity: Lessons from Mouse Models. Front. Immunol. 2018, 9, 104. [Google Scholar] [CrossRef]
- Ando, M.; Sato, Y.; Takata, K.; Nomoto, J.; Nakamura, S.; Ohshima, K.; Takeuchi, T.; Orita, Y.; Kobayashi, Y.; Yoshino, T. A20 (TNFAIP3) deletion in Epstein-Barr virus-associated lymphoproliferative disorders/lymphomas. PLoS ONE 2013, 8, e56741. [Google Scholar] [CrossRef]
- Malakhova, O.A.; Kim, K.I.; Luo, J.K.; Zou, W.; Kumar, K.G.; Fuchs, S.Y.; Shuai, K.; Zhang, D.E. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 2006, 25, 2358–2367. [Google Scholar] [CrossRef]
- Le, J.; Perez, E.; Nemzow, L.; Gong, F. Role of deubiquitinases in DNA damage response. DNA Repair 2019, 76, 89–98. [Google Scholar] [CrossRef] [PubMed]
- Lai, K.P.; Chen, J.; Tse, W.K.F. Role of Deubiquitinases in Human Cancers: Potential Targeted Therapy. Int. J. Mol. Sci. 2020, 21, 2548. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, N.; Hirose, S. Regulation of mitochondrial morphology by USP30, a deubiquitinating enzyme present in the mitochondrial outer membrane. Mol. Biol. Cell 2008, 19, 1903–1911. [Google Scholar] [CrossRef]
- Wauer, T.; Swatek, K.N.; Wagstaff, J.L.; Gladkova, C.; Pruneda, J.N.; Michel, M.A.; Gersch, M.; Johnson, C.M.; Freund, S.M.; Komander, D. Ubiquitin Ser65 phosphorylation affects ubiquitin structure, chain assembly and hydrolysis. EMBO J. 2015, 34, 307–325. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.N.; Baughman, J.M.; Phu, L.; Tea, J.S.; Yu, C.; Coons, M.; Kirkpatrick, D.S.; Bingol, B.; Corn, J.E. USP30 and parkin homeostatically regulate atypical ubiquitin chains on mitochondria. Nat. Cell Biol. 2015, 17, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Bingol, B.; Tea, J.S.; Phu, L.; Reichelt, M.; Bakalarski, C.E.; Song, Q.; Foreman, O.; Kirkpatrick, D.S.; Sheng, M. The mitochondrial deubiquitinase USP30 opposes parkin-mediated mitophagy. Nature 2014, 510, 370–375. [Google Scholar] [CrossRef]
- Sato, Y.; Yoshikawa, A.; Yamagata, A.; Mimura, H.; Yamashita, M.; Ookata, K.; Nureki, O.; Iwai, K.; Komada, M.; Fukai, S. Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 2008, 455, 358–362. [Google Scholar] [CrossRef]
- Wang, D.; Xu, C.; Yang, W.; Chen, J.; Ou, Y.; Guan, Y.; Guan, J.; Liu, Y. E3 ligase RNF167 and deubiquitinase STAMBPL1 modulate mTOR and cancer progression. Mol. Cell 2022, 82, 770–784.e9. [Google Scholar] [CrossRef]
- Mahul-Mellier, A.L.; Pazarentzos, E.; Datler, C.; Iwasawa, R.; AbuAli, G.; Lin, B.; Grimm, S. De-ubiquitinating protease USP2a targets RIP1 and TRAF2 to mediate cell death by TNF. Cell Death Differ. 2012, 19, 891–899. [Google Scholar] [CrossRef]
- Xu, G.; Tan, X.; Wang, H.; Sun, W.; Shi, Y.; Burlingame, S.; Gu, X.; Cao, G.; Zhang, T.; Qin, J.; et al. Ubiquitin-specific peptidase 21 inhibits tumor necrosis factor α-induced nuclear factor κB activation via binding to and deubiquitinating receptor-interacting protein 1. J. Biol. Chem. 2010, 285, 969–978. [Google Scholar] [CrossRef]
- Goricke, F.; Vu, V.; Smith, L.; Scheib, U.; Bohm, R.; Akkilic, N.; Wohlfahrt, G.; Weiske, J.; Bomer, U.; Brzezinka, K.; et al. Discovery and Characterization of BAY-805, a Potent and Selective Inhibitor of Ubiquitin-Specific Protease USP21. J. Med. Chem. 2023, 66, 3431–3447. [Google Scholar] [CrossRef] [PubMed]
- Pajtler, K.W.; Mack, S.C.; Ramaswamy, V.; Smith, C.A.; Witt, H.; Smith, A.; Hansford, J.R.; von Hoff, K.; Wright, K.D.; Hwang, E.; et al. The current consensus on the clinical management of intracranial ependymoma and its distinct molecular variants. Acta Neuropathol. 2017, 133, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Hyrskyluoto, A.; Bruelle, C.; Lundh, S.H.; Do, H.T.; Kivinen, J.; Rappou, E.; Reijonen, S.; Waltimo, T.; Petersen, A.; Lindholm, D.; et al. Ubiquitin-specific protease-14 reduces cellular aggregates and protects against mutant huntingtin-induced cell degeneration: Involvement of the proteasome and ER stress-activated kinase IRE1α. Hum. Mol. Genet. 2014, 23, 5928–5939. [Google Scholar] [CrossRef]
- Giordano, M.; Roncagalli, R.; Bourdely, P.; Chasson, L.; Buferne, M.; Yamasaki, S.; Beyaert, R.; van Loo, G.; Auphan-Anezin, N.; Schmitt-Verhulst, A.M.; et al. The tumor necrosis factor alpha-induced protein 3 (TNFAIP3, A20) imposes a brake on antitumor activity of CD8 T cells. Proc. Natl. Acad. Sci. USA 2014, 111, 11115–11120. [Google Scholar] [CrossRef] [PubMed]
- Momtazi, G.; Lambrecht, B.N.; Naranjo, J.R.; Schock, B.C. Regulators of A20 (TNFAIP3): New drug-able targets in inflammation. Am. J. Physiol. Lung Cell. Mol. Physiol. 2019, 316, L456–L469. [Google Scholar] [CrossRef]
- van Bon, B.W.M.; Mefford, H.C.; de Vries, B.B.A.; Schaaf, C.P. 15q13.3 Recurrent Deletion. In GeneReviews((R)); Adam, M.P., Mirzaa, G.M., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Gripp, K.W., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Uddin, M.; Unda, B.K.; Kwan, V.; Holzapfel, N.T.; White, S.H.; Chalil, L.; Woodbury-Smith, M.; Ho, K.S.; Harward, E.; Murtaza, N.; et al. OTUD7A Regulates Neurodevelopmental Phenotypes in the 15q13.3 Microdeletion Syndrome. Am. J. Hum. Genet. 2018, 102, 278–295. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.H.; Ha, U.H.; Woo, C.H.; Xu, H.; Li, J.D. CYLD is a crucial negative regulator of innate immune response in Escherichia coli pneumonia. Cell. Microbiol. 2008, 10, 2247–2256. [Google Scholar] [CrossRef]
- Deng, M.; Dai, W.; Yu, V.Z.; Tao, L.; Lung, M.L. Cylindromatosis Lysine 63 Deubiquitinase (CYLD) Regulates NF-kB Signaling Pathway and Modulates Fibroblast and Endothelial Cells Recruitment in Nasopharyngeal Carcinoma. Cancers 2020, 12, 1924. [Google Scholar] [CrossRef]
- Sun, S.C. CYLD: A tumor suppressor deubiquitinase regulating NF-κB activation and diverse biological processes. Cell Death Differ. 2010, 17, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, L.F.; Sparks, A.; Allende-Vega, N.; Xirodimas, D.P.; Lane, D.P.; Saville, M.K. The deubiquitinating enzyme USP2a regulates the p53 pathway by targeting Mdm2. EMBO J. 2007, 26, 976–986. [Google Scholar] [CrossRef] [PubMed]
- Shan, J.; Zhao, W.; Gu, W. Suppression of cancer cell growth by promoting cyclin D1 degradation. Mol. Cell 2009, 36, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Duguay, D.; Fahrenkrug, J.; Cermakian, N.; Wing, S.S. USP2 regulates the intracellular localization of PER1 and circadian gene expression. J. Biol. Rhythm. 2014, 29, 243–256. [Google Scholar] [CrossRef] [PubMed]
- Scoma, H.D.; Humby, M.; Yadav, G.; Zhang, Q.; Fogerty, J.; Besharse, J.C. The de-ubiquitinylating enzyme, USP2, is associated with the circadian clockwork and regulates its sensitivity to light. PLoS ONE 2011, 6, e25382. [Google Scholar] [CrossRef]
- Stojkovic, K.; Wing, S.S.; Cermakian, N. A central role for ubiquitination within a circadian clock protein modification code. Front. Mol. Neurosci. 2014, 7, 69. [Google Scholar] [CrossRef]
- He, J.; Lee, H.J.; Saha, S.; Ruan, D.; Guo, H.; Chan, C.H. Inhibition of USP2 eliminates cancer stem cells and enhances TNBC responsiveness to chemotherapy. Cell Death Dis. 2019, 10, 285. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, J.; Wang, X.; Cai, S.; Guo, Y.; Ye, L.; Li, D.; Hu, A.; Jin, S.; Yuan, B.; et al. Therapeutic targeting of the USP2-E2F4 axis inhibits autophagic machinery essential for zinc homeostasis in cancer progression. Autophagy 2022, 18, 2615–2635. [Google Scholar] [CrossRef]
- Lee, J.H.; Zou, L.; Yang, R.; Han, J.; Wan, Q.; Zhang, X.; El Baghdady, S.; Roman, A.; Elly, C.; Jin, H.S.; et al. The deubiquitinase CYLD controls protective immunity against helminth infection by regulation of Treg cell plasticity. J. Allergy Clin. Immunol. 2021, 148, 209–224.e9. [Google Scholar] [CrossRef]
- Bustamante, H.A.; Cereceda, K.; Gonzalez, A.E.; Valenzuela, G.E.; Cheuquemilla, Y.; Hernandez, S.; Arias-Munoz, E.; Cerda-Troncoso, C.; Bandau, S.; Soza, A.; et al. The Proteasomal Deubiquitinating Enzyme PSMD14 Regulates Macroautophagy by Controlling Golgi-to-ER Retrograde Transport. Cells 2020, 9, 777. [Google Scholar] [CrossRef]
- Bustamante, H.A.; Albornoz, N.; Morselli, E.; Soza, A.; Burgos, P.V. Novel insights into the non-canonical roles of PSMD14/POH1/Rpn11 in proteostasis and in the modulation of cancer progression. Cell. Signal. 2023, 101, 110490. [Google Scholar] [CrossRef]
- Zhang, D.; Zaugg, K.; Mak, T.W.; Elledge, S.J. A role for the deubiquitinating enzyme USP28 in control of the DNA-damage response. Cell 2006, 126, 529–542. [Google Scholar] [CrossRef]
- Jacq, X.; Kemp, M.; Martin, N.M.; Jackson, S.P. Deubiquitylating enzymes and DNA damage response pathways. Cell Biochem. Biophys. 2013, 67, 25–43. [Google Scholar] [CrossRef]
- Liang, X.W.; Wang, S.Z.; Liu, B.; Chen, J.C.; Cao, Z.; Chu, F.R.; Lin, X.; Liu, H.; Wu, J.C. A review of deubiquitinases and thier roles in tumorigenesis and development. Front. Bioeng. Biotechnol. 2023, 11, 1204472. [Google Scholar] [CrossRef]
- Huo, Y.; Khatri, N.; Hou, Q.; Gilbert, J.; Wang, G.; Man, H.Y. The deubiquitinating enzyme USP46 regulates AMPA receptor ubiquitination and trafficking. J. Neurochem. 2015, 134, 1067–1080. [Google Scholar] [CrossRef] [PubMed]
- Anggono, V.; Huganir, R.L. Regulation of AMPA receptor trafficking and synaptic plasticity. Curr. Opin. Neurobiol. 2012, 22, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Ishiuchi, S.; Tsuzuki, K.; Yoshida, Y.; Yamada, N.; Hagimura, N.; Okado, H.; Miwa, A.; Kurihara, H.; Nakazato, Y.; Tamura, M.; et al. Blockage of Ca(2+)-permeable AMPA receptors suppresses migration and induces apoptosis in human glioblastoma cells. Nat. Med. 2002, 8, 971–978. [Google Scholar] [CrossRef] [PubMed]
- Piao, Y.; Lu, L.; de Groot, J. AMPA receptors promote perivascular glioma invasion via β1 integrin-dependent adhesion to the extracellular matrix. Neuro. Oncol. 2009, 11, 260–273. [Google Scholar] [CrossRef]
- Krishna, S.; Choudhury, A.; Keough, M.B.; Seo, K.; Ni, L.; Kakaizada, S.; Lee, A.; Aabedi, A.; Popova, G.; Lipkin, B.; et al. Glioblastoma remodelling of human neural circuits decreases survival. Nature 2023, 617, 599–607. [Google Scholar] [CrossRef]
- Tomida, S.; Mamiya, T.; Sakamaki, H.; Miura, M.; Aosaki, T.; Masuda, M.; Niwa, M.; Kameyama, T.; Kobayashi, J.; Iwaki, Y.; et al. Usp46 is a quantitative trait gene regulating mouse immobile behavior in the tail suspension and forced swimming tests. Nat. Genet. 2009, 41, 688–695. [Google Scholar] [CrossRef]
- Xian, J.; Zhang, Q.; Guo, X.; Liang, X.; Liu, X.; Feng, Y. A prognostic signature based on three non-coding RNAs for prediction of the overall survival of glioma patients. FEBS Open Bio. 2019, 9, 682–692. [Google Scholar] [CrossRef] [PubMed]
- Smits, A.; Jin, Z.; Elsir, T.; Pedder, H.; Nister, M.; Alafuzoff, I.; Dimberg, A.; Edqvist, P.H.; Ponten, F.; Aronica, E.; et al. GABA-A channel subunit expression in human glioma correlates with tumor histology and clinical outcome. PLoS ONE 2012, 7, e37041. [Google Scholar] [CrossRef] [PubMed]
- Blanchart, A.; Fernando, R.; Haring, M.; Assaife-Lopes, N.; Romanov, R.A.; Andang, M.; Harkany, T.; Ernfors, P. Endogenous GAB(AA) receptor activity suppresses glioma growth. Oncogene 2017, 36, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Lin, K.; Chang, G.; Chen, Y.; Yue, C.; Guo, Q.; Zhang, S.; Jia, Z.; Huang, T.T.; Zhou, A.; et al. Aberrant Activation of β-Catenin Signaling Drives Glioma Tumorigenesis via USP1-Mediated Stabilization of EZH2. Cancer Res. 2019, 79, 72–85. [Google Scholar] [CrossRef]
- Li, Z.; Li, M.; Wang, D.; Hou, P.; Chen, X.; Chu, S.; Chai, D.; Zheng, J.; Bai, J. Post-translational modifications of EZH2 in cancer. Cell Biosci. 2020, 10, 143. [Google Scholar] [CrossRef]
- Nibe, Y.; Oshima, S.; Kobayashi, M.; Maeyashiki, C.; Matsuzawa, Y.; Otsubo, K.; Matsuda, H.; Aonuma, E.; Nemoto, Y.; Nagaishi, T.; et al. Novel polyubiquitin imaging system, PolyUb-FC, reveals that K33-linked polyubiquitin is recruited by SQSTM1/p62. Autophagy 2018, 14, 347–358. [Google Scholar] [CrossRef]
- Chen, Y.H.; Chen, H.H.; Wang, W.J.; Chen, H.Y.; Huang, W.S.; Kao, C.H.; Lee, S.R.; Yeat, N.Y.; Yan, R.L.; Chan, S.J.; et al. TRABID inhibition activates cGAS/STING-mediated anti-tumor immunity through mitosis and autophagy dysregulation. Nat. Commun. 2023, 14, 3050. [Google Scholar] [CrossRef]
- Afonina, I.S.; Beyaert, R. Trabid epigenetically drives expression of IL-12 and IL-23. Nat. Immunol. 2016, 17, 227–228. [Google Scholar] [CrossRef]
- Jin, J.; Xie, X.; Xiao, Y.; Hu, H.; Zou, Q.; Cheng, X.; Sun, S.C. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat. Immunol. 2016, 17, 259–268. [Google Scholar] [CrossRef]
- Ma, J.; Zhou, Y.; Pan, P.; Yu, H.; Wang, Z.; Li, L.L.; Wang, B.; Yan, Y.; Pan, Y.; Ye, Q.; et al. TRABID overexpression enables synthetic lethality to PARP inhibitor via prolonging 53BP1 retention at double-strand breaks. Nat. Commun. 2023, 14, 1810. [Google Scholar] [CrossRef]
- Wertz, I.E.; O’Rourke, K.M.; Zhou, H.; Eby, M.; Aravind, L.; Seshagiri, S.; Wu, P.; Wiesmann, C.; Baker, R.; Boone, D.L.; et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature 2004, 430, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Wertz, I.E.; Newton, K.; Seshasayee, D.; Kusam, S.; Lam, C.; Zhang, J.; Popovych, N.; Helgason, E.; Schoeffler, A.; Jeet, S.; et al. Phosphorylation and linear ubiquitin direct A20 inhibition of inflammation. Nature 2015, 528, 370–375. [Google Scholar] [CrossRef] [PubMed]
- Abbasi, A.; Forsberg, K.; Bischof, F. The role of the ubiquitin-editing enzyme A20 in diseases of the central nervous system and other pathological processes. Front. Mol. Neurosci. 2015, 8, 21. [Google Scholar] [CrossRef] [PubMed]
- Catrysse, L.; Vereecke, L.; Beyaert, R.; van Loo, G. A20 in inflammation and autoimmunity. Trends Immunol. 2014, 35, 22–31. [Google Scholar] [CrossRef]
- Opipari, A.W., Jr.; Boguski, M.S.; Dixit, V.M. The A20 cDNA induced by tumor necrosis factor alpha encodes a novel type of zinc finger protein. J. Biol. Chem. 1990, 265, 14705–14708. [Google Scholar] [CrossRef]
- Bredel, M.; Bredel, C.; Juric, D.; Duran, G.E.; Yu, R.X.; Harsh, G.R.; Vogel, H.; Recht, L.D.; Scheck, A.C.; Sikic, B.I. Tumor necrosis factor-alpha-induced protein 3 as a putative regulator of nuclear factor-κB-mediated resistance to O6-alkylating agents in human glioblastomas. J. Clin. Oncol. 2006, 24, 274–287. [Google Scholar] [CrossRef]
- Hjelmeland, A.B.; Wu, Q.; Wickman, S.; Eyler, C.; Heddleston, J.; Shi, Q.; Lathia, J.D.; Macswords, J.; Lee, J.; McLendon, R.E.; et al. Targeting A20 decreases glioma stem cell survival and tumor growth. PLoS Biol. 2010, 8, e1000319. [Google Scholar] [CrossRef]
- Sharma, T.; Schwalbe, E.C.; Williamson, D.; Sill, M.; Hovestadt, V.; Mynarek, M.; Rutkowski, S.; Robinson, G.W.; Gajjar, A.; Cavalli, F.; et al. Second-generation molecular subgrouping of medulloblastoma: An international meta-analysis of Group 3 and Group 4 subtypes. Acta Neuropathol. 2019, 138, 309–326. [Google Scholar] [CrossRef]
- Jung, M.; Russell, A.J.; Liu, B.; George, J.; Liu, P.Y.; Liu, T.; DeFazio, A.; Bowtell, D.D.; Oberthuer, A.; London, W.B.; et al. A Myc Activity Signature Predicts Poor Clinical Outcomes in Myc-Associated Cancers. Cancer Res. 2017, 77, 971–981. [Google Scholar] [CrossRef]
- Kitamura, H.; Hashimoto, M. USP2-Related Cellular Signaling and Consequent Pathophysiological Outcomes. Int. J. Mol. Sci. 2021, 22, 1209. [Google Scholar] [CrossRef]
- Benassi, B.; Flavin, R.; Marchionni, L.; Zanata, S.; Pan, Y.; Chowdhury, D.; Marani, M.; Strano, S.; Muti, P.; Blandino, G.; et al. MYC is activated by USP2a-mediated modulation of microRNAs in prostate cancer. Cancer Discov. 2012, 2, 236–247. [Google Scholar] [CrossRef]
- Petrosyan, E.; Fares, J.; Fernandez, L.G.; Yeeravalli, R.; Dmello, C.; Duffy, J.T.; Zhang, P.; Lee-Chang, C.; Miska, J.; Ahmed, A.U.; et al. Endoplasmic Reticulum Stress in the Brain Tumor Immune Microenvironment. Mol. Cancer Res. 2023, 21, 389–396. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, D.; Shireman, J.; Sierra Potchanant, E.A.; Lara-Velazquez, M.; Dey, M. Neuroinflammation in Autoimmune Disease and Primary Brain Tumors: The Quest for Striking the Right Balance. Front. Cell. Neurosci. 2021, 15, 716947. [Google Scholar] [CrossRef] [PubMed]
- Pelizzari-Raymundo, D.; Doultsinos, D.; Pineau, R.; Sauzay, C.; Koutsandreas, T.; Langlais, T.; Carlesso, A.; Gkotsi, E.; Negroni, L.; Avril, T.; et al. A novel IRE1 kinase inhibitor for adjuvant glioblastoma treatment. iScience 2023, 26, 106687. [Google Scholar] [CrossRef] [PubMed]
- Flores-Santibanez, F.; Rennen, S.; Fernandez, D.; De Nolf, C.; Van De Velde, E.; Gaete Gonzalez, S.; Fuentes, C.; Moreno, C.; Figueroa, D.; Lladser, A.; et al. Nuanced role for dendritic cell intrinsic IRE1 RNase in the regulation of antitumor adaptive immunity. Front. Immunol. 2023, 14, 1209588. [Google Scholar] [CrossRef]
- Liang, W.; Fang, J.; Zhou, S.; Hu, W.; Yang, Z.; Li, Z.; Dai, L.; Tao, Y.; Fu, X.; Wang, X. The role of ubiquitin-specific peptidases in glioma progression. Biomed. Pharmacother. 2022, 146, 112585. [Google Scholar] [CrossRef]
- Hanna, J.; Hathaway, N.A.; Tone, Y.; Crosas, B.; Elsasser, S.; Kirkpatrick, D.S.; Leggett, D.S.; Gygi, S.P.; King, R.W.; Finley, D. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell 2006, 127, 99–111. [Google Scholar] [CrossRef]
- Lee, B.H.; Lee, M.J.; Park, S.; Oh, D.C.; Elsasser, S.; Chen, P.C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar] [CrossRef]
- Lee, J.H.; Shin, S.K.; Jiang, Y.; Choi, W.H.; Hong, C.; Kim, D.E.; Lee, M.J. Facilitated Tau Degradation by USP14 Aptamers via Enhanced Proteasome Activity. Sci. Rep. 2015, 5, 10757. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhang, C.; Gu, C.; Li, Q.; Wu, N. Function of Deubiquitinating Enzyme USP14 as Oncogene in Different Types of Cancer. Cell. Physiol. Biochem. 2016, 38, 993–1002. [Google Scholar] [CrossRef]
- Ma, Y.S.; Wang, X.F.; Zhang, Y.J.; Luo, P.; Long, H.D.; Li, L.; Yang, H.Q.; Xie, R.T.; Jia, C.Y.; Lu, G.X.; et al. Inhibition of USP14 Deubiquitinating Activity as a Potential Therapy for Tumors with p53 Deficiency. Mol. Ther. Oncolytics 2020, 16, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Kaokhum, N.; Pinto-Fernandez, A.; Wilkinson, M.; Kessler, B.M.; Ismail, H.M. The Mechano-Ubiquitinome of Articular Cartilage: Differential Ubiquitination and Activation of a Group of ER-Associated DUBs and ER Stress Regulators. Mol. Cell. Proteom. 2022, 21, 100419. [Google Scholar] [CrossRef]
- Ernst, R.; Mueller, B.; Ploegh, H.L.; Schlieker, C. The otubain YOD1 is a deubiquitinating enzyme that associates with p97 to facilitate protein dislocation from the ER. Mol. Cell 2009, 36, 28–38. [Google Scholar] [CrossRef] [PubMed]
- Schimmack, G.; Schorpp, K.; Kutzner, K.; Gehring, T.; Brenke, J.K.; Hadian, K.; Krappmann, D. YOD1/TRAF6 association balances p62-dependent IL-1 signaling to NF-κB. Elife 2017, 6, e22416. [Google Scholar] [CrossRef]
- Tanji, K.; Mori, F.; Miki, Y.; Utsumi, J.; Sasaki, H.; Kakita, A.; Takahashi, H.; Wakabayashi, K. YOD1 attenuates neurogenic proteotoxicity through its deubiquitinating activity. Neurobiol. Dis. 2018, 112, 14–23. [Google Scholar] [CrossRef]
- Zhou, L.; Li, L.; Chen, Y.; Chen, C.; Zhi, Z.; Yan, L.; Wang, Y.; Liu, B.; Zhai, Q. miR-190a-3p Promotes Proliferation and Migration in Glioma Cells via YOD1. Comput. Math. Methods Med. 2021, 2021, 3957738. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Kim, W.; Song, Y.; Kim, J.R.; Cho, K.; Moon, H.; Ro, S.W.; Seo, E.; Ryu, Y.M.; Myung, S.J.; et al. Deubiquitinase YOD1 potentiates YAP/TAZ activities through enhancing ITCH stability. Proc. Natl. Acad. Sci. USA 2017, 114, 4691–4696. [Google Scholar] [CrossRef] [PubMed]
- Masliantsev, K.; Karayan-Tapon, L.; Guichet, P.O. Hippo Signaling Pathway in Gliomas. Cells 2021, 10, 184. [Google Scholar] [CrossRef]
- Nijman, S.M.; Huang, T.T.; Dirac, A.M.; Brummelkamp, T.R.; Kerkhoven, R.M.; D’Andrea, A.D.; Bernards, R. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 2005, 17, 331–339. [Google Scholar] [CrossRef]
- Huang, T.T.; Nijman, S.M.; Mirchandani, K.D.; Galardy, P.J.; Cohn, M.A.; Haas, W.; Gygi, S.P.; Ploegh, H.L.; Bernards, R.; D’Andrea, A.D. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 2006, 8, 339–347. [Google Scholar] [CrossRef]
- Kashiwaba, S.; Kanao, R.; Masuda, Y.; Kusumoto-Matsuo, R.; Hanaoka, F.; Masutani, C. USP7 Is a Suppressor of PCNA Ubiquitination and Oxidative-Stress-Induced Mutagenesis in Human Cells. Cell Rep. 2015, 13, 2072–2080. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.L.; Dianova, I.I.; Khoronenkova, S.V.; Edelmann, M.J.; Kessler, B.M.; Dianov, G.L. USP47 is a deubiquitylating enzyme that regulates base excision repair by controlling steady-state levels of DNA polymerase β. Mol. Cell 2011, 41, 609–615. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Majid, M.C.; Soll, J.M.; Brickner, J.R.; Dango, S.; Mosammaparast, N. Noncanonical regulation of alkylation damage resistance by the OTUD4 deubiquitinase. EMBO J. 2015, 34, 1687–1703. [Google Scholar] [CrossRef]
- Chitale, S.; Richly, H. Timing of DNA lesion recognition: Ubiquitin signaling in the NER pathway. Cell Cycle 2017, 16, 163–171. [Google Scholar] [CrossRef]
- Perez-Oliva, A.B.; Lachaud, C.; Szyniarowski, P.; Munoz, I.; Macartney, T.; Hickson, I.; Rouse, J.; Alessi, D.R. USP45 deubiquitylase controls ERCC1-XPF endonuclease-mediated DNA damage responses. EMBO J. 2015, 34, 326–343. [Google Scholar] [CrossRef]
- Nishi, R.; Wijnhoven, P.; le Sage, C.; Tjeertes, J.; Galanty, Y.; Forment, J.V.; Clague, M.J.; Urbe, S.; Jackson, S.P. Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 2014, 16, 1016–1026. [Google Scholar] [CrossRef]
- Sato, Y.; Yamagata, A.; Goto-Ito, S.; Kubota, K.; Miyamoto, R.; Nakada, S.; Fukai, S. Molecular basis of Lys-63-linked polyubiquitination inhibition by the interaction between human deubiquitinating enzyme OTUB1 and ubiquitin-conjugating enzyme UBC13. J. Biol. Chem. 2012, 287, 25860–25868. [Google Scholar] [CrossRef] [PubMed]
- Butler, L.R.; Densham, R.M.; Jia, J.; Garvin, A.J.; Stone, H.R.; Shah, V.; Weekes, D.; Festy, F.; Beesley, J.; Morris, J.R. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 2012, 31, 3918–3934. [Google Scholar] [CrossRef]
- Kakarougkas, A.; Ismail, A.; Katsuki, Y.; Freire, R.; Shibata, A.; Jeggo, P.A. Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res. 2013, 41, 10298–10311. [Google Scholar] [CrossRef]
- Vincent, P.; Collette, Y.; Marignier, R.; Vuaillat, C.; Rogemond, V.; Davoust, N.; Malcus, C.; Cavagna, S.; Gessain, A.; Machuca-Gayet, I.; et al. A role for the neuronal protein collapsin response mediator protein 2 in T lymphocyte polarization and migration. J. Immunol. 2005, 175, 7650–7660. [Google Scholar] [CrossRef]
- Nakamura, F.; Ohshima, T.; Goshima, Y. Collapsin Response Mediator Proteins: Their Biological Functions and Pathophysiology in Neuronal Development and Regeneration. Front. Cell. Neurosci. 2020, 14, 188. [Google Scholar] [CrossRef]
- Bedekovics, T.; Hussain, S.; Zhang, Y.; Ali, A.; Jeon, Y.J.; Galardy, P.J. USP24 Is a Cancer-Associated Ubiquitin Hydrolase, Novel Tumor Suppressor, and Chromosome Instability Gene Deleted in Neuroblastoma. Cancer Res. 2021, 81, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Thayer, J.A.; Awad, O.; Hegdekar, N.; Sarkar, C.; Tesfay, H.; Burt, C.; Zeng, X.; Feldman, R.A.; Lipinski, M.M. The PARK10 gene USP24 is a negative regulator of autophagy and ULK1 protein stability. Autophagy 2020, 16, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Qi, W.; Yang, F.; Pan, H. Deubiquitinase JOSD1 promotes tumor progression via stabilizing Snail in lung adenocarcinoma. Am. J. Cancer Res. 2022, 12, 2323–2336. [Google Scholar]
- Yang, J.; Weisberg, E.L.; Liu, X.; Magin, R.S.; Chan, W.C.; Hu, B.; Schauer, N.J.; Zhang, S.; Lamberto, I.; Doherty, L.; et al. Small molecule inhibition of deubiquitinating enzyme JOSD1 as a novel targeted therapy for leukemias with mutant JAK2. Leukemia 2022, 36, 210–220. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Zhang, Y.; Zhao, P.; Qian, L.; Yuan, Y.; Liu, J.; Cheng, Q.; Xu, W.; Zuo, Y.; et al. JOSD1 Negatively Regulates Type-I Interferon Antiviral Activity by Deubiquitinating and Stabilizing SOCS1. Viral Immunol. 2017, 30, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Zhou, J.; Tang, J.; Zhou, F.; He, Z.; Liu, T.; Liu, T. MINDY1 promotes bladder cancer progression by stabilizing YAP. Cancer Cell Int. 2021, 21, 395. [Google Scholar] [CrossRef]
- Tang, J.; Luo, Y.; Long, G.; Zhou, L. MINDY1 promotes breast cancer cell proliferation by stabilizing estrogen receptor α. Cell Death Dis. 2021, 12, 937. [Google Scholar] [CrossRef] [PubMed]













| Astrocytoma 1 | DUB | p Value (Corrected) | Chromosome | Versus Non-Tumor Group in the Sun Dataset |
|---|---|---|---|---|
| CYLD | Usp | 2.44 × 10−13 | 16 | Down |
| USP12 | Usp | 3.73 × 10−12 | 13 | Down |
| USP3 | Usp | 1.95 × 10−11 | 15 | Up |
| OTUD7A | Otu | 1.26 × 10−10 | 15 | Down |
| OTUB1 | Otu | 4.38 × 10−10 | 11 | Down |
| USP8 | Usp | 1.69 × 10−09 | 15 | Up |
| USP33 | Usp | 8.97 × 10−09 | 1 | Down |
| STAMBPL1 | Jamm | 5.64 × 10−08 | 10 | Down |
| EIF3F | Jamm | 5.94 × 10−08 | 11 | Up |
| EIF3H | Jamm | 4.60 × 10−07 | 8 | Up |
| Glioblastoma 2 | ||||
| USP12 | Usp | 5.22 × 10−20 | 13 | Down |
| OTUD7A | Otu | 1.13 × 10−19 | 15 | Down |
| ZRANB1 | Otu | 2.37 × 10−18 | 10 | Down |
| USP46 | Usp | 3.60 × 10−18 | 4 | Down |
| OTUB1 | Otu | 4.24 × 10−18 | 11 | Down |
| CYLD | Usp | 2.34 × 10−16 | 16 | Down |
| USP3 | Usp | 1.20 × 10−13 | 15 | Up |
| USP27X | Usp | 5.35 × 10−12 | X | Down |
| USP30 | Usp | 9.20 × 10−12 | 12 | Down |
| USP11 | Usp | 1.39 × 10−11 | X | Down |
| Oligodendroglioma 3 | ||||
| USP12 | Usp | 3.27 × 10−11 | 13 | Down |
| OTUD7A | Otu | 3.55 × 10−10 | 15 | Down |
| USP48 | Usp | 6.67 × 10−09 | 1 | Down |
| USP33 | Usp | 1.37 × 10−08 | 1 | Down |
| CYLD | Usp | 1.45 × 10−08 | 16 | Down |
| USP3 | Usp | 1.50 × 10−08 | 15 | Up |
| EIF3F | Jamm | 1.57 × 10−08 | 11 | Up |
| OTUD3 | Otu | 2.46 × 10−08 | 1 | Down |
| USP14 | Usp | 4.85 × 10−08 | 18 | Down |
| OTUB1 | Otu | 6.68 × 10−08 | 11 | Down |
| DUB Gene | Family | Chromosome | Chi Square Kaplan Meier | p Value | Hazard Ratio (HR) | HR p Value | Better Survival |
|---|---|---|---|---|---|---|---|
| ZRANB1 | Otu | 10 | 116.69 | 3.36 × 10−27 | 0.21 | 4.6 × 10−27 | High |
| FAM188A | Mindy | 10 | 68.95 | 1.01 × 10−16 | 0.53 | 1.4 × 10−13 | High |
| USP34 | Usp | 2 | 63.01 | 2.06 × 10−15 | 0.30 | 4.4 × 10−12 | High |
| USP49 | Usp | 6 | 63.46 | 1.63 × 10−15 | 0.55 | 1.9 × 10−16 | High |
| Usp27X | Usp | X | 57.58 | 3.24 × 10−14 | 0.43 | 1.6 × 10−15 | High |
| USP54 | Usp | 10 | 51.46 | 7.30 × 10−13 | 0.68 | 2.5 × 10−11 | High |
| USP51 | Usp | X | 49.10 | 2.43 × 10−12 | 0.69 | 1.4 × 10−11 | High |
| USP30 | Usp | 12 | 43.77 | 3.69 × 10−11 | 0.47 | 1.5 × 10−08 | High |
| USP11 | Usp | X | 42.18 | 8.33 × 10−11 | 0.52 | 2.6 × 10−10 | High |
| OTUD7A | Otu | 15 | 38.15 | 6.54 × 10−10 | 0.69 | 1.3 × 10−09 | High |
| EIF3H | Jamm | 8 | 35.16 | 3.40 × 10−09 | 0.47 | 7.1 × 10−10 | High |
| USP1 | Usp | 1 | 33.71 | 6.39 × 10−09 | 1.8 | 1.4 × 10−08 | Low |
| USP4 | Usp | 3 | 33.07 | 8.90 × 10−09 | 2.6 | 5.6 × 10−08 | Low |
| ATXN3 | MJD | 14 | 33.10 | 8.75 × 10−09 | 0.65 | 7.7 × 10−06 | High |
| USP43 | Usp | 17 | 31.76 | 1.74 × 10−08 | 0.74 | 1.1 × 10−08 | High |
| USP46 | Usp | 4 | 31.45 | 2.05 × 10−08 | 0.65 | 9.2 × 10−08 | High |
| TNFAIP3 | Otu | 6 | 30.86 | 2.77 × 10−08 | 1.3 | 6.0 × 10−07 | Low |
| OTUD1 | Otu | 10 | 28.27 | 1.06 × 10−07 | 0.54 | 2.0 × 10−08 | High |
| JOSD2 | MJD | 19 | 25.88 | 3.62 × 10−07 | 1.5 | 2.1 × 10−04 | Low |
| PSMD7 | Jamm | 16 | 24.22 | 8.60 × 10−07 | 1.9 | 1.3 × 10−04 | Low |
| STAMBPL1 | Jamm | 10 | 23.87 | 1.03 × 10−06 | 0.76 | 4.3 × 10−05 | High |
| USP14 | Usp | 18 | 23.95 | 9.90 × 10−07 | 2.1 | 1.0 × 10−04 | low |
| DUB Gene | Non-Tumor N = 23 | Astrocytoma N = 26 | Glioblastoma N = 77 | Oligodendroglioma N = 50 |
|---|---|---|---|---|
| USP14 | 851.87 ± 25.31 | 689.66 ± 25.63 * | 738.5 ± 18.13 * | 647.71 ± 16.28 * |
| USP19 | 159.11 ± 6.47 | 206.93 ± 9.5 * | 222.11 ± 6.09 * | 226.03 ± 7.9 * |
| USP25 | 314.57 ± 14.19 | 216.77 ± 9.34 * | 216.88 ± 9.18 * | 209.77 ± 7.59 * |
| BAP1 | 295.14 ± 10.78 | 221.5 ± 7.97 * | 244.66 ± 5.02 * | 248.25 ± 6.88 * |
| Group 3 (N = 233) | DUB | p Value vs. Non-Tumor | Chromosome | Versus Non-Tumor Group in the Swartling Dataset |
|---|---|---|---|---|
| USP46 | Usp | 1.16 × 10−57 | 4 | Down |
| USP2 | Usp | 5.71 × 10−53 | 11 | Up |
| PSMD14 | Jamm | 4.25 × 10−51 | 2 | Up |
| USP49 | Usp | 1.34 × 10−36 | 6 | Up |
| USP28 | Usp | 2.41 × 10−26 | 11 | Down |
| USP30 | Usp | 1.29 × 10−25 | 12 | Up |
| UCHL1 | Uch | 1.72 × 10−22 | 4 | Down |
| OTUD7A | Otu | 6.20 × 10−22 | 15 | Down |
| USP44 | Usp | 6.39 × 10−22 | 12 | Down |
| COPS6 | Jamm | 2.19 × 10−19 | 7 | Up |
| Group 4 (N = 530) | ||||
| USP20 | Usp | 8.96 × 10−65 | 9 | Up |
| USP28 | Usp | 2.28 × 10−63 | 11 | Down |
| USP22 | Usp | 1.35 × 10−61 | 17 | Up |
| USP32 | Usp | 1.21 × 10−58 | 17 | Up |
| USP3 | Usp | 2.98 × 10−54 | 15 | Down |
| USP30 | Usp | 2.19 × 10−52 | 12 | Up |
| USP49 | Usp | 1.49 × 10−43 | 6 | Up |
| USP45 | Usp | 9.56 × 10−36 | 6 | Down |
| USP36 | Usp | 7.95 × 10−29 | 17 | Up |
| EIF3H | Jamm | 1.59 × 10−27 | 8 | Down |
| SHH (N = 405) | ||||
| EIF3H | Jamm | 4.51 × 19−135 | 8 | Up |
| USP2 | Usp | 1.17 × 10−88 | 11 | Down |
| USP20 | Usp | 2.26 × 10−69 | 9 | Down |
| CYLD | Usp | 7.13 × 10−64 | 16 | Down |
| USP25 | Usp | 4.63 × 10−40 | 21 | Down |
| USP32 | Usp | 4.97 × 10−38 | 17 | Down |
| USP33 | Usp | 5.42 × 10−36 | 1 | Down |
| JOSD1 | Mjd | 2.40 × 10−30 | 22 | Up |
| USP11 | Usp | 3.32 × 10−28 | X | Down |
| USP13 | Usp | 1.03 × 10−26 | 3 | Up |
| WNT (N = 118) | ||||
| UCHL1 | Uch | 2.14 × 10−82 | 4 | Down |
| USP2 | Usp | 3.18 × 10−56 | 11 | Down |
| USP20 | Usp | 6.88 × 10−50 | 9 | Down |
| USP32 | Usp | 2.23 × 10−47 | 17 | Down |
| USP5 | Usp | 1.19 × 10−38 | 12 | Up |
| COPS6 | Jamm | 2.59 × 10−32 | 7 | Up |
| EIF3H | Jamm | 1.19 × 10−29 | 8 | Up |
| OTUD7A | Otu | 1.48 × 10−29 | 15 | Down |
| USP33 | Usp | 1.38 × 10−28 | 1 | Down |
| USP28 | Usp | 6.05 × 10−27 | 11 | Down |
| DUB | DUB Family | Chromosome | Chi Square Kaplan Meier | p Value | Hazard Ratio | p Value for Hazard Ratio | Better Survival |
|---|---|---|---|---|---|---|---|
| VCPIP1 | Otu | 8 | 28.14 | 1.13 × 10−07 | 1.9 | 1.10 × 10−05 | Low |
| USP49 | Usp | 6 | 26.73 | 2.34 × 10−07 | 1.9 | 1.30 × 10−05 | Low |
| USP2 | Usp | 11 | 21.82 | 2.99 × 10−06 | 1.3 | 3.50 × 10−05 | Low |
| USP51 | Usp | X | 19.36 | 1.08 × 10−05 | 0.44 | 1.00 × 10−04 | High |
| STAMBPL1 | Jamm | 10 | 16.70 | 4.37 × 10−05 | 0.73 | 3.10 × 10−04 | High |
| PSMD14 | Jamm | 2 | 18.05 | 2.16 × 19−05 | 1.4 | 4.20 × 10−04 | Low |
| ZRANB1 | Otu | 10 | 17.11 | 3.52 × 10−05 | 0.54 | 1.00 × 10−03 | High |
| OTUD3 | Otu | 1 | 18.76 | 1.48 × 10−05 | 2.4 | 1.30 × 10−03 | Low |
| PRPF8 | Jamm | 17 | 18.77 | 1.47 × 10−05 | 0.58 | 2.20 × 10−03 | High |
| USP15 | Usp | 12 | 16.42 | 5.06 × 10−05 | 2 | 3.00 × 10−03 | Low |
| USP45 | Usp | 6 | 14.82 | 1.18 × 10−04 | 1.5 | 3.90 × 10−03 | Low |
| USP26 | Usp | X | 22.77 | 1.82 × 10−06 | 7.3 | 5.20 × 10−03 | Low |
| USP36 | Usp | 17 | 22.07 | 2.63 × 10−06 | 2.2 | 5.50 × 10−03 | Low |
| USPL1 | Usp | 13 | 14.89 | 1.14 × 10−04 | 2.1 | 5.80 × 10−03 | Low |
| USP25 | Usp | 21 | 12.36 | 4.39 × 10−04 | 1.6 | 8.70 × 10−03 | Low |
| EIF3H | Jamm | 8 | 13.23 | 2.75 × 10−04 | 1.5 | 9.40 × 10−03 | Low |
| COPS5 | Jamm | 8 | 9.25 | 2.36 × 10−04 | 1.9 | 9.90 × 10−03 | Low |
| DUB | Family | p Value Corrected FDR | Versus Non-Tumor Group in Swartling Dataset |
|---|---|---|---|
| Group 3 (n = 233) | |||
| PSMD14 | Jamm | 8.74 × 10−52 | Up |
| OTUD7A | Otu | 1.70 × 10−22 | Down |
| TNFAIP3 | Otu | 6.26 × 10−10 | Up |
| CYLD | Usp | 3.54 × 10−08 | Down |
| Group 4 (n = 530) | |||
| PSMD14 | Jamm | 2.23 × 10−26 | Up |
| OTUD7A | Otu | 1.16 × 10−13 | Down |
| CYLD | Usp | 7.04 × 10−10 | Up |
| TNFAIP3 | Otu | 5.67 × 10−09 | Up |
| SHH (n = 405) | |||
| CYLD | Usp | 1.95 × 10−64 | Down |
| PSMD14 | Jamm | 4.95 × 10−09 | Down |
| TNFAIP3 | Otu | 2.95 × 10−08 | Up |
| PSMD7 | Jamm | 5.19 × 10−08 | Up |
| Wnt (n = 118) | |||
| OTUD7A | Otu | 8.10 × 10−30 | Down |
| PSMD7 | Jamm | 3.89 × 10−25 | Up |
| TNFAIP3 | Otu | 2.40 × 10−10 | Down |
| CYLD | Usp | 4.19 × 10−04 | Up |
| PSMD14 | Jamm | 8.99 × 10−04 | Up |
| DUB | Family | p Value vs. NT Group Swartling | Versus Non-Tumor Group in Swartling Dataset |
|---|---|---|---|
| Group 3 vs. NT | |||
| PSMD14 | Jamm | 2.27 × 10−51 | Up |
| USP28 | Usp | 1.07 × 10−26 | Down |
| COPS6 | Jamm | 1.30 × 10−21 | Up |
| USP47 | Usp | 3.96 × 10−15 | Down |
| UCHL5 | Uch | 6.12 × 10−14 | Up |
| COPS5 | Jamm | 1.61 × 10−10 | Up |
| USP1 | Usp | 2.53 × 10−06 | Up |
| OTUB1 | Otu | 3.33 × 10−05 | Down |
| Group 4 vs. NT | |||
| Usp28 | Usp | 8.12 × 10−64 | Down |
| USP3 | Usp | 1.33 × 10−54 | Down |
| USP45 | Usp | 4.54 × 10−36 | Down |
| PSMD14 | Jamm | 1.45 × 10−26 | Up |
| COPS6 | Jamm | 3.10 × 10−24 | Up |
| USP1 | Usp | 1.48 × 10−16 | Up |
| OTUB1 | Otu | 1.67 × 10−14 | Down |
| USP7 | Usp | 2.65 × 10−12 | Up |
| COPS5 | Jamm | 4.01 × 10−06 | Down |
| USP47 | Usp | 2.16 × 10−04 | Down |
| UCHL5 | Uch | 2.11 × 10−03 | Up |
| SHH vs. NT | |||
| USP10 | Usp | 2.21 × 10−15 | Up |
| PSMD14 | Jamm | 8.58 × 10−09 | Down |
| COPS5 | Jamm | 1.16 × 10−08 | Up |
| USP45 | Usp | 3.60 × 10−07 | Down |
| UCHL5 | Uch | 1.30 × 10−06 | Down |
| USP3 | Usp | 5.90 × 10−06 | Down |
| COPS6 | Jamm | 5.98 × 10−06 | Down |
| USP1 | Usp | 6.06 × 10−06 | Up |
| USP47 | Usp | 6.68 × 10−04 | Down |
| USP7 | Usp | 1.71 × 10−03 | Up |
| WNT vs. NT | |||
| COPS6 | Jamm | 2.67 × 10−32 | Up |
| USP28 | Usp | 5.39 × 10−27 | Down |
| USP3 | Usp | 3.53 × 10−25 | Down |
| USP45 | Usp | 2.61 × 10−22 | Down |
| UCHL5 | Usp | 4.72 × 10−14 | Down |
| USP10 | Usp | 7.73 × 10−11 | Up |
| COPS5 | Jamm | 1.02 × 10−03 | Up |
| PSMD14 | Jamm | 1.46 × 10−03 | Up |
| OTUB1 | Otu | 2.89 × 10−03 | Up |
| USP1 | Usp | 4.55 × 10−03 | Up |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klonisch, T.; Logue, S.E.; Hombach-Klonisch, S.; Vriend, J. DUBing Primary Tumors of the Central Nervous System: Regulatory Roles of Deubiquitinases. Biomolecules 2023, 13, 1503. https://doi.org/10.3390/biom13101503
Klonisch T, Logue SE, Hombach-Klonisch S, Vriend J. DUBing Primary Tumors of the Central Nervous System: Regulatory Roles of Deubiquitinases. Biomolecules. 2023; 13(10):1503. https://doi.org/10.3390/biom13101503
Chicago/Turabian StyleKlonisch, Thomas, Susan E. Logue, Sabine Hombach-Klonisch, and Jerry Vriend. 2023. "DUBing Primary Tumors of the Central Nervous System: Regulatory Roles of Deubiquitinases" Biomolecules 13, no. 10: 1503. https://doi.org/10.3390/biom13101503
APA StyleKlonisch, T., Logue, S. E., Hombach-Klonisch, S., & Vriend, J. (2023). DUBing Primary Tumors of the Central Nervous System: Regulatory Roles of Deubiquitinases. Biomolecules, 13(10), 1503. https://doi.org/10.3390/biom13101503