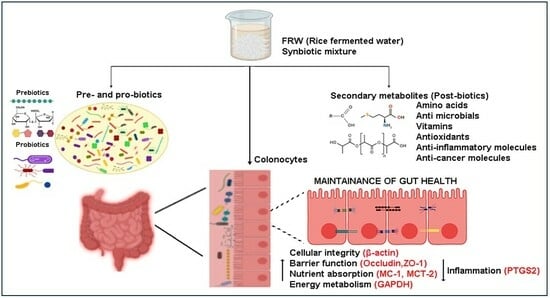
Postbiotics of Naturally Fermented Synbiotic Mixture of Rice Water Aids in Promoting Colonocyte Health
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals, Reagents, and Growth Media
2.2. Preparation of Fermented Rice Water (FRW) and Non-Fermented Rice Water (nFRW) from the South Indian Rice Varieties
2.3. Proximate Composition Analysis
2.4. Microbial Culture for Isolation, Enumeration and Identification of the Microbial Species Grown in FRW
2.5. Lyophilisation of Fermented Samples
2.6. Sample Preparation for Metabolomic Analysis
2.7. Data Acquisition
2.7.1. UHPLC
2.7.2. MS
2.8. Reconstitution of the Fermented Rice Water
2.9. Cell Culture
2.10. Cell Viability Assay
2.11. mRNA Isolation and cDNA Conversion
2.12. Gene Expression Studies Using qPCR
2.13. Statistical Analysis
3. Results
3.1. Natural Fermentation of Edible Rice Enrich for Probiotics
3.1.1. Rice-Based Carbon Source Alone Is Enough for Short-Duration Natural Fermentation
3.1.2. Natural Fermentation of Edible Rice Enables Growth of Probiotics of the LAB Group
3.1.3. Environmental Influence toward Natural Fermentation of Edible Rice
3.2. Macronutrient Analysis of Each Fermented Rice Variety and Its Water
3.3. Metabolomic Analysis of the WP-Fermented Rice Water
3.4. Functional Analysis of FRW in the Colonocyte Cell Line HT29
3.4.1. FRW from Different Rice Varieties Differentially Modulate Proliferation of HT29 Cells
3.4.2. Probiotics Generated Biomolecules Aid in Improving Colonocyte Health
Postbiotics in FRW Upregulate Nutrient Absorptive Genes
Fermented Rice Water Modulates the Expression of Cytoskeletal Genes
Fermented Rice Water Modulates the Metabolic Pathway
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Konkel, L. The environment within: Exploring the role of the gut microbiome in health and disease. Environ. Health Perspect. 2013, 121, A276–A281. [Google Scholar] [CrossRef]
- Marchesi, J.R.; Adams, D.H.; Fava, F.; Hermes, G.D.; Hirschfield, G.M.; Hold, G.; Quraishi, M.N.; Kinross, J.; Smidt, H.; Tuohy, K.M.; et al. The gut microbiota and host health: A new clinical frontier. Gut 2016, 65, 330–339. [Google Scholar] [CrossRef]
- Bosi, A.; Banfi, D.; Bistoletti, M.; Giaroni, C.; Baj, A. Tryptophan Metabolites along the Microbiota-Gut-Brain Axis: An Interkingdom Communication System Influencing the Gut in Health and Disease. Int. J. Tryptophan Res. 2020, 13, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, N.S.; Lorentz, A. Dietary restrictions modulate the gut microbiota: Implications for health and disease. Nutr. Res. 2021, 89, 10–22. [Google Scholar] [CrossRef]
- Milani, C.; Duranti, S.; Bottacini, F.; Casey, E.; Turroni, F.; Mahony, J.; Belzer, C.; Delgado Palacio, S.; Arboleya Montes, S.; Mancabelli, L.; et al. The First Microbial Colonizers of the Human Gut: Composition, Activities, and Health Implications of the Infant Gut Microbiota. Microbiol. Mol. Biol. Rev. 2017, 81, e00036-17. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef] [PubMed]
- Danneskiold-Samsoe, N.B.; Dias de Freitas Queiroz Barros, H.; Santos, R.; Bicas, J.L.; Cazarin, C.B.B.; Madsen, L.; Kristiansen, K.; Pastore, G.M.; Brix, S.; Marostica Junior, M.R. Interplay between food and gut microbiota in health and disease. Food Res. Int. 2019, 115, 23–31. [Google Scholar] [CrossRef]
- Wong, J.M.; de Souza, R.; Kendall, C.W.; Emam, A.; Jenkins, D.J. Colonic health: Fermentation and short chain fatty acids. J. Clin. Gastroenterol. 2006, 40, 235–243. [Google Scholar] [CrossRef]
- Dahiya, D.; Nigam, P.S. Antibiotic-Therapy-Induced Gut Dysbiosis Affecting Gut Microbiota-Brain Axis and Cognition: Restoration by Intake of Probiotics and Synbiotics. Int. J. Mol. Sci. 2023, 24, 3074. [Google Scholar] [CrossRef]
- Bellucci, E.; Chiereghin, F.; Pacifici, F.; Donadel, G.; De Stefano, A.; Malatesta, G.; Valente, M.G.; Guadagni, F.; Infante, M.; Rovella, V.; et al. Novel therapeutic approaches based on the pathological role of gut dysbiosis on the link between nonalcoholic fatty liver disease and insulin resistance. Eur. Rev. Med. Pharmacol. Sci. 2023, 27, 1921–1944. [Google Scholar] [CrossRef]
- Tongthong, T.; Kaewduangduen, W.; Phuengmaung, P.; Chancharoenthana, W.; Leelahavanichkul, A. Lacticaseibacillus rhamnosus dfa1 Attenuate Cecal Ligation-Induced Systemic Inflammation through the Interference in Gut Dysbiosis, Leaky Gut, and Enterocytic Cell Energy. Int. J. Mol. Sci. 2023, 24, 3756. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Shi, Y.; Qiu, W.; Lin, Q.; Zeng, S.; Hou, Y.; Zhou, H.; Chen, M.; Zhang, D. Limosilactobacillus reuteri ameliorates preeclampsia in mice via improving gut dysbiosis and endothelial dysfunction. Biomed. Pharmacother. 2023, 161, 114429. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Wang, J.; Du, L.; Chen, J.; Zheng, Q.; Li, P.; Du, B.; Fang, X.; Liao, Z. Kefir microbiota and metabolites stimulate intestinal mucosal immunity and its early development. Crit. Rev. Food Sci. Nutr. 2022, 64, 1371–1384. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.; Kang, X.; Yang, H.; Liu, H.; Yang, X.; Liu, Q.; Tian, H.; Xue, Y.; Ren, P.; Kuang, X.; et al. Lactobacillus acidophilus ameliorates obesity in mice through modulation of gut microbiota dysbiosis and intestinal permeability. Pharmacol. Res. 2022, 175, 106020. [Google Scholar] [CrossRef] [PubMed]
- Bidell, M.R.; Hobbs, A.L.V.; Lodise, T.P. Gut microbiome health and dysbiosis: A clinical primer. Pharmacotherapy 2022, 42, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Wasser, C.I.; Mercieca, E.C.; Kong, G.; Hannan, A.J.; Allford, B.; McKeown, S.J.; Stout, J.C.; Glikmann-Johnston, Y. A Randomized Controlled Trial of Probiotics Targeting Gut Dysbiosis in Huntington’s Disease. J. Huntingt. Dis. 2023, 12, 43–55. [Google Scholar] [CrossRef]
- Xie, L.; Alam, M.J.; Marques, F.Z.; Mackay, C.R. A major mechanism for immunomodulation: Dietary fibres and acid metabolites. Semin. Immunol. 2023, 66, 101737. [Google Scholar] [CrossRef]
- Kumar, J.; Rani, K.; Datt, C. Molecular link between dietary fibre, gut microbiota and health. Mol. Biol. Rep. 2020, 47, 6229–6237. [Google Scholar] [CrossRef]
- Meng, H.Y.H.; Mak, C.C.H.; Mak, W.Y.; Zuo, T.; Ko, H.; Chan, F.K.L. Probiotic supplementation demonstrates therapeutic potential in treating gut dysbiosis and improving neurocognitive function in age-related dementia. Eur. J. Nutr. 2022, 61, 1701–1734. [Google Scholar] [CrossRef]
- Yoon, W.; Park, S.H.; Lee, J.S.; Byeon, J.H.; Kim, S.H.; Lim, J.; Yoo, Y. Probiotic mixture reduces gut inflammation and microbial dysbiosis in children with atopic dermatitis. Australas. J. Dermatol. 2021, 62, e386–e392. [Google Scholar] [CrossRef] [PubMed]
- Haarhuis, J.E.; Kardinaal, A.; Kortman, G.A.M. Probiotics, prebiotics and postbiotics for better sleep quality: A narrative review. Benef. Microbes 2022, 13, 169–182. [Google Scholar] [CrossRef]
- Scarpellini, E.; Rinninella, E.; Basilico, M.; Colomier, E.; Rasetti, C.; Larussa, T.; Santori, P.; Abenavoli, L. From Pre- and Probiotics to Post-Biotics: A Narrative Review. Int. J. Environ. Res. Public Health 2021, 19, 37. [Google Scholar] [CrossRef]
- Yang, K.; Wang, X.; Huang, R.; Wang, H.; Lan, P.; Zhao, Y. Prebiotics and Postbiotics Synergistic Delivery Microcapsules from Microfluidics for Treating Colitis. Adv. Sci. 2022, 9, e2104089. [Google Scholar] [CrossRef] [PubMed]
- Jastrzab, R.; Graczyk, D.; Siedlecki, P. Molecular and Cellular Mechanisms Influenced by Postbiotics. Int. J. Mol. Sci. 2021, 22, 13475. [Google Scholar] [CrossRef]
- Salminen, S.; Collado, M.C.; Endo, A.; Hill, C.; Lebeer, S.; Quigley, E.M.M.; Sanders, M.E.; Shamir, R.; Swann, J.R.; Szajewska, H.; et al. The International Scientific Association of Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 649–667. [Google Scholar] [CrossRef]
- Scott, E.; De Paepe, K.; Van de Wiele, T. Postbiotics and Their Health Modulatory Biomolecules. Biomolecules 2022, 12, 1640. [Google Scholar] [CrossRef] [PubMed]
- Kavita; Om, H.; Chand, U.; Kushawaha, P.K. Postbiotics: An alternative and innovative intervention for the therapy of inflammatory bowel disease. Microbiol. Res. 2024, 279, 127550. [Google Scholar] [CrossRef]
- Rad, A.H.; Aghebati-Maleki, L.; Kafil, H.S.; Abbasi, A. Molecular mechanisms of postbiotics in colorectal cancer prevention and treatment. Crit. Rev. Food Sci. Nutr. 2021, 61, 1787–1803. [Google Scholar] [CrossRef] [PubMed]
- Cabello-Olmo, M.; Arana, M.; Urtasun, R.; Encio, I.J.; Barajas, M. Role of Postbiotics in Diabetes Mellitus: Current Knowledge and Future Perspectives. Foods 2021, 10, 1590. [Google Scholar] [CrossRef]
- Kolonics, A.; Bori, Z.; Torma, F.; Abraham, D.; Feher, J.; Radak, Z. Exercise combined with postbiotics treatment results in synergistic improvement of mitochondrial function in the brain of male transgenic mice for Alzheimer’s disease. BMC Neurosci. 2023, 24, 68. [Google Scholar] [CrossRef]
- Park, M.; Joung, M.; Park, J.H.; Ha, S.K.; Park, H.Y. Role of Postbiotics in Diet-Induced Metabolic Disorders. Nutrients 2022, 14, 3701. [Google Scholar] [CrossRef]
- Han, S.; Seo, K.H.; Gyu Lee, H.; Kim, H. Effect of Cucumis melo L. peel extract supplemented postbiotics on reprograming gut microbiota and sarcopenia in hindlimb-immobilized mice. Food Res. Int. 2023, 173 Pt 2, 113476. [Google Scholar] [CrossRef] [PubMed]
- Tomova, A.; Bukovsky, I.; Rembert, E.; Yonas, W.; Alwarith, J.; Barnard, N.D.; Kahleova, H. The Effects of Vegetarian and Vegan Diets on Gut Microbiota. Front. Nutr. 2019, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Mercado-Mercado, G.; Blancas-Benitez, F.J.; Zamora-Gasga, V.M.; Sayago-Ayerdi, S.G. Mexican Traditional Plant-Foods: Polyphenols Bioavailability, Gut Microbiota Metabolism and Impact Human Health. Curr. Pharm. Des. 2019, 25, 3434–3456. [Google Scholar] [CrossRef] [PubMed]
- Ozma, M.A.; Abbasi, A.; Akrami, S.; Lahouty, M.; Shahbazi, N.; Ganbarov, K.; Pagliano, P.; Sabahi, S.; Kose, S.; Yousefi, M.; et al. Postbiotics as the key mediators of the gut microbiota-host interactions. Infez. Med. 2022, 30, 180–193. [Google Scholar] [CrossRef]
- Warda, A.K.; Clooney, A.G.; Ryan, F.; de Almeida Bettio, P.H.; Di Benedetto, G.; Ross, R.P.; Hill, C. A postbiotic consisting of heat-treated lactobacilli has a bifidogenic effect in pure culture and in human fermented faecal communities. Appl. Environ. Microbiol. 2021, 87, e02459-20. [Google Scholar] [CrossRef] [PubMed]
- Youn, H.Y.; Seo, K.H.; Kim, H.J.; Kim, Y.S.; Kim, H. Effect of postbiotics derived from kefir lactic acid bacteria-mediated bioconversion of citrus pomace extract and whey on high-fat diet-induced obesity and gut dysbiosis. Food Res. Int. 2022, 162 Pt A, 111930. [Google Scholar] [CrossRef]
- Mayta-Apaza, A.C.; Pottgen, E.; De Bodt, J.; Papp, N.; Marasini, D.; Howard, L.; Abranko, L.; Van de Wiele, T.; Lee, S.O.; Carbonero, F. Impact of tart cherries polyphenols on the human gut microbiota and phenolic metabolites in vitro and in vivo. J. Nutr. Biochem. 2018, 59, 160–172. [Google Scholar] [CrossRef]
- Ismaeel, A.; Valentino, T.R.; Burke, B.; Goh, J.; Saliu, T.P.; Albathi, F.; Owen, A.; McCarthy, J.J.; Wen, Y. Acetate and succinate benefit host muscle energetics as exercise-associated post-biotics. Physiol. Rep. 2023, 11, e15848. [Google Scholar] [CrossRef]
- Sangeetha, K.N.; Vennila, R.; Secunda, R.; Sakthivel, S.; Pathak, S.; Jeswanth, S.; Surendran, R. Functional variations between Mesenchymal Stem Cells of different tissue origins: A comparative gene expression profiling. Biotechnol. Lett. 2020, 42, 1287–1304. [Google Scholar] [CrossRef]
- Koirala, P.; Maina, N.H.; Nihtila, H.; Katina, K.; Coda, R. Brewers’ spent grain as substrate for dextran biosynthesis by Leuconostoc pseudomesenteroides DSM20193 and Weissella confusa A16. Microb. Cell Fact. 2021, 20, 23. [Google Scholar] [CrossRef]
- Lee, M.A.; Choi, Y.J.; Kim, Y.S.; Chon, S.Y.; Chung, Y.B.; Park, S.H.; Yun, Y.R.; Min, S.G.; Yang, H.C.; Seo, H.Y. Effects of salt type on the metabolites and microbial community in kimchi fermentation. Heliyon 2022, 8, e11360. [Google Scholar] [CrossRef]
- Dan, H.; Gu, Z.; Li, C.; Fang, Z.; Hu, B.; Wang, C.; Chen, S.; Tang, X.; Ren, Y.; Wu, W.; et al. Effect of fermentation time and addition amount of rice sourdoughs with different microbial compositions on the physicochemical properties of three gluten-free rice breads. Food Res. Int. 2022, 161, 111889. [Google Scholar] [CrossRef]
- Bhatia, R.; Sharma, S.; Bhadada, S.K.; Bishnoi, M.; Kondepudi, K.K. Lactic Acid Bacterial Supplementation Ameliorated the Lipopolysaccharide-Induced Gut Inflammation and Dysbiosis in Mice. Front. Microbiol. 2022, 13, 930928. [Google Scholar] [CrossRef]
- Wang, G.; Si, Q.; Yang, S.; Jiao, T.; Zhu, H.; Tian, P.; Wang, L.; Li, X.; Gong, L.; Zhao, J.; et al. Lactic acid bacteria reduce diabetes symptoms in mice by alleviating gut microbiota dysbiosis and inflammation in different manners. Food Funct. 2020, 11, 5898–5914. [Google Scholar] [CrossRef]
- Hung, Y.P.; Lee, C.C.; Lee, J.C.; Tsai, P.J.; Ko, W.C. Gut Dysbiosis during COVID-19 and Potential Effect of Probiotics. Microorganisms 2021, 9, 1605. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Nie, C.; Zhao, J.; Ou, X.; Zhang, Y.; Yang, S.; Bai, X.; Wang, Y.; Wang, J.; Li, J. Microbiome-Metabolomics Analysis of the Impacts of Long-Term Dietary Advanced-Glycation-End-Product Consumption on C57BL/6 Mouse Fecal Microbiota and Metabolites. J. Agric. Food Chem. 2018, 66, 8864–8875. [Google Scholar] [CrossRef] [PubMed]
- Verbeke, K.A.; Boobis, A.R.; Chiodini, A.; Edwards, C.A.; Franck, A.; Kleerebezem, M.; Nauta, A.; Raes, J.; van Tol, E.A.; Tuohy, K.M. Towards microbial fermentation metabolites as markers for health benefits of prebiotics. Nutr. Res. Rev. 2015, 28, 42–66. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Zaiss, M.M.; Jones, R.M.; Schett, G.; Pacifici, R. The gut-bone axis: How bacterial metabolites bridge the distance. J. Clin. Investig. 2019, 129, 3018–3028. [Google Scholar] [CrossRef]
- Dinu, L.D.; Gatea, F.; Roaming Israel, F.; Lakicevic, M.; Dedovic, N.; Vamanu, E. The Modulation Effect of a Fermented Bee Pollen Postbiotic on Cardiovascular Microbiota and Therapeutic Perspectives. Biomedicines 2023, 11, 2712. [Google Scholar] [CrossRef] [PubMed]
- Duan, W.; Zhou, L.; Ren, Y.; Liu, F.; Xue, Y.; Wang, F.Z.; Lu, R.; Zhang, X.J.; Shi, J.S.; Xu, Z.H.; et al. Lactic acid fermentation of goji berries (Lycium barbarum) prevents acute alcohol liver injury and modulates gut microbiota and metabolites in mice. Food Funct. 2024, 15, 1612–1626. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yan, C.; Wang, L.; Ai, C.; Wang, S.; Shen, C.; Tong, Y.; Song, S. Ascophyllum nodosum polysaccharide regulates gut microbiota metabolites to protect against colonic inflammation in mice. Food Funct. 2023, 14, 810–821. [Google Scholar] [CrossRef] [PubMed]
- Daliri, E.B. Untargeted Metabolomics of Fermented Rice Using UHPLC Q-TOF MS/MS Reveals an Abundance of Potential Antihypertensive Compounds. Foods 2020, 9, 8. [Google Scholar] [CrossRef]







| S. No | Rice Variety | Location | Probiotic Bacteria Identified | CFU/mL |
|---|---|---|---|---|
| 1. | Raw Rice | Location 2 | Lactococcus lactis ssp. lactis | 54 × 109 |
| Weisella confusa | ||||
| Location 4 | Leuconostoc lactis | 47.4 × 106 | ||
| 2. | White Ponni | Location 3 (over two-nights fermented) | Lactococcus lactis | 85.5 × 109 |
| Lactobacillus nagelii | ||||
| Lactobacillus delbrueckii ssp. indicus | ||||
| Location 3 (overnight fermented) | Leuconostoc lactis | UD | ||
| Lactobacillus fermentum | ||||
| Location 1 | Leuconostoc lactis | 15 × 108 | ||
| Weisella confusa | 50 × 106 | |||
| Location 4 | lactococcus taiwanensis | 1.28 × 105 | ||
| Lactococcus lactis | ||||
| 3. | Mappillai Samba # | Location 4 | Leuconostoc lactis | 6.3 × 108 |
| Weisella cibacria | ||||
| Weisella confusa | ||||
| 4. | Control * | Location 4 | Lactobacillus plantarum | UD |
| S. No | Rice Variety | Location | Other Bacteria |
|---|---|---|---|
| 1. | Raw rice | Location 4 | Streptococcus equinus |
| Acinetobacter baumannii | |||
| Franconibacter pulveris | |||
| 2 | White Ponni | Location 1 | Acinetobacter baumannii |
| Streptococcus lutetiensis | |||
| Sterptococcus infantaurius | |||
| Kocuria kristinae | |||
| Location 4 | Franconibacter pulveris | ||
| Enterobacter cloacae | |||
| Acinetobacter baumannii | |||
| Cronobacter sp. | |||
| 3 | Mappillai Samba | Location 4 | Klebsiella penumoniae ssp. pneumoniae |
| Franconibacter pulveris | |||
| Bacillus cereus | |||
| Enterococcus gallinarum | |||
| Sterptococcus infantaurius ssp. infantarius | |||
| Enterobacter cloacae complex | |||
| 4 | Control | Location 4 | Enterococcus faecium |
| S. No | Rice Varieties | Rice/ Rice Water | Types of Fermentation | Moisture (%) | Dry Matter Basis (%) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Crude Protein | Crude Fibre | Ether Extract | Total Ash | Nitrogen Free Extract (NFE) | |||||
| 1. | Raw rice * | Rice | Before fermentation | 80.9 | 8.55 | 0.16 | 0.22 | 0.28 | 90.79 |
| Non-fermentation | 80.63 | 8.10 | 0.18 | 0.31 | 0.26 | 91.15 | |||
| After fermentation | 79.9 | 8.6 | 0.19 | 0.38 | 0.26 | 90.57 | |||
| Rice water | Before fermentation | 95.69 | 6.51 | Not traceable | 0.29 | 0.46 | 92.74 | ||
| Non-fermentation | 96.44 | 6.88 | Not traceable | 0.14 | 0.72 | 92.26 | |||
| After fermentation | 96.0 | 5.98 | Not traceable | 0.14 | 0.62 | 93.26 | |||
| 2. | Mappillai samba | Rice | Before fermentation | 80.45 | 9.18 | 0.48 | 0.98 | 0.58 | 88.78 |
| Non-fermentation | 85.18 | 8.5 | 0.52 | 1.04 | 0.55 | 89.39 | |||
| After fermentation | 85.42 | 8.87 | 0.6 | 1.25 | 0.47 | 88.81 | |||
| Rice Water | Before fermentation | 98.19 | 4.21 | Not traceable | 0.34 | 1.45 | 94.0 | ||
| Non-fermentation | 98.27 | 4.45 | Not traceable | 0.20 | 1.59 | 93.76 | |||
| After fermentation | 97.4 | 4.77 | Not traceable | 0.19 | 1.61 | 93.43 | |||
| 3. | Parboiled rice * | Rice | Before fermentation | 81.96 | 8.62 | 0.31 | 0.50 | 0.46 | 90.11 |
| Non-fermentation | 80.19 | 8.59 | 0.46 | 0.46 | 0.54 | 89.95 | |||
| After fermentation | 81.5 | 9.05 | 0.28 | 0.58 | 0.50 | 89.59 | |||
| Rice Water | Before fermentation | 97.02 | 6.01 | 0.21 | 0.36 | 1.81 | 91.61 | ||
| Non-fermentation | 99.03 | 7.30 | 0.25 | 0.54 | 2.72 | 89.19 | |||
| After fermentation | 96.73 | 4.97 | Not traceable | 0.28 | 1.78 | 92.97 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Anbalagan, C.; Nandabalan, S.K.; Sankar, P.; Rajaram, P.S.; Govindaraj, K.; Rupert, S.; Sathyanesan, J. Postbiotics of Naturally Fermented Synbiotic Mixture of Rice Water Aids in Promoting Colonocyte Health. Biomolecules 2024, 14, 344. https://doi.org/10.3390/biom14030344
Anbalagan C, Nandabalan SK, Sankar P, Rajaram PS, Govindaraj K, Rupert S, Sathyanesan J. Postbiotics of Naturally Fermented Synbiotic Mixture of Rice Water Aids in Promoting Colonocyte Health. Biomolecules. 2024; 14(3):344. https://doi.org/10.3390/biom14030344
Chicago/Turabian StyleAnbalagan, Charumathi, Sangeetha Kadapakkam Nandabalan, Pavithra Sankar, Prasanna Srinivasan Rajaram, Karthick Govindaraj, Secunda Rupert, and Jeswanth Sathyanesan. 2024. "Postbiotics of Naturally Fermented Synbiotic Mixture of Rice Water Aids in Promoting Colonocyte Health" Biomolecules 14, no. 3: 344. https://doi.org/10.3390/biom14030344
APA StyleAnbalagan, C., Nandabalan, S. K., Sankar, P., Rajaram, P. S., Govindaraj, K., Rupert, S., & Sathyanesan, J. (2024). Postbiotics of Naturally Fermented Synbiotic Mixture of Rice Water Aids in Promoting Colonocyte Health. Biomolecules, 14(3), 344. https://doi.org/10.3390/biom14030344