Amyloid Fibrils from Hemoglobin
Abstract
:1. Introduction
2. Results
2.1. Amyloid Fibril Formation from Waste Blood Hemoglobin
2.2. Characterisation of AHB Nanofibers
2.2.1. Thioflavin T Fluorescence
2.2.2. Circular Dichroism
2.2.3. TEM Imaging
2.2.4. AFM Imaging and Analysis
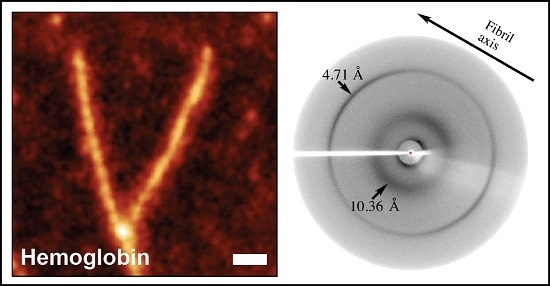
2.2.5. X-ray Fiber Diffraction
2.3. Stability of AHB Fibrils
2.3.1. Solvent Stability of AHB Fibrils
2.3.2. Temperature Stability of AHB Fibrils
2.3.3. AHB Fibril Stability under a Range of pHs
2.3.4. Protease Resistance of AHB Fibrils
2.4. Scaled up Formation of AHB Fibrils from Bovine Waste Blood
3. Discussion
3.1. Amyloid Fibril Formation from Waste Blood Hemoglobin
3.2. Characterisation of AHB Nanofibers
3.3. Stability of AHB Fibrils
3.3.1. Solvent Stability of AHB Fibrils
3.3.2. Temperature Stability of AHB Fibrils
3.3.3. AHB Fibril Stability under a Range of pHs
3.3.4. Protease Resistance of AHB Fibrils
3.4. Scaled up Formation of AHB Fibrils from Bovine Waste Blood
4. Materials and Methods
4.1. Materials
4.2. Preparation of Apo-Hemoglobin from Bovine Blood
4.3. LC–MS Analysis of AHB Solutions
4.4. Fibril Formation from AHB
4.5. ThT Assay
4.6. Circular Dichroism
4.7. AFM
4.8. TEM
4.9. X-ray Fiber Diffraction
4.10. Stability of AHB Fibrils
4.11. Large Scale Fibril Preparation
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Chiti, F.; Dobson, M. Protein misfolding, functional amyloid, and human disease. Annu. Rev. Biochem. 2006, 75, 333–366. [Google Scholar] [CrossRef] [PubMed]
- Hughes, V.A.; Dunstan, D.E. Amyloid fibrils - self-assembling proteins. Mod. Polym. Sci. 2009, 559–594. [Google Scholar]
- Xue, W-F.; Homans, S.W.; Radford, S.E. Systematic analysis of nucleation-dependent polymerization reveals new insights into the mechanism of amyloid self-assembly. Proc. Natl. Acad. Sci. USA 2008, 105, 8926–8931. [Google Scholar]
- Fändrich, M.; Fletcher, M.A.; Dobson, C.M. Amyloid fibrils from muscle myoglobin. Nature 2001, 410, 165–166. [Google Scholar] [CrossRef] [PubMed]
- Dobson, C.M. Protein folding and misfolding. Nature 2003, 426, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.F.; Knowles, T.P.; Dobson, C.M.; Macphee, C.E.; Welland, M.E. Characterization of the nanoscale properties of individual amyloid fibrils. Proc. Natl. Acad. Sci. USA 2006, 103, 15806–15811. [Google Scholar] [CrossRef] [PubMed]
- Shammas, S.L.; Knowles, T.P.J.; Baldwin, A.J.; Macphee, C.E.; Welland, M.E.; Dobson, C.M.; Devlin, G.L. Perturbation of the stability of amyloid fibrils through alteration of electrostatic interactions. Biophys. J. 2011, 100, 2783–2791. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Mezzenga, R. Amyloid fibrils as building blocks for natural and artificial functional materials. Adv. Mater. 2016, 28, 6546–6561. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Adamcik, J.; Mezzenga, R. Biodegradable nanocomposties of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nat. Nanotechnol. 2012, 7, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Domigan, L.; Andersen, K.B.; Sasso, L.; Dimaki, M.; Svendsen, W.E.; Gerrard, J.A.; Castillo-León, J. Dielectrophoretic manipulation and solubility of protein nanofibrils formed from crude crystallins. Electrophoresis 2013, 34, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Sasso, L.; Suei, S.; Domigan, L.; Healy, J.; Nock, V.; Williams, M.A.K.; Gerrard, J.A. Versatile multi-functionalization of protein nanofibrils for biosensor applications. Nanoscale 2014, 6, 1629–1634. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Roberts, S.; Healy, J.; Domigan, L.; Vasudevamurthy, M.; Gerrard, J.A.; Sasso, L. Crystallin nanofibrils: A functionalizable nanoscaffold with broad applications manufactured from waste. Chempluschem 2015, 80, 810–819. [Google Scholar] [CrossRef]
- Sasso, L.; Gerrard, J.A. Self-Assembled Biological Nanofibers for Biosensor Applications. In Micro and Nanofabrication Using Self-Assembled Biological Nanostructures, 1st ed.; Elsevier Inc.: Amsterdam, Netharlands, 2015; pp. 1–20. [Google Scholar]
- Scheibel, T.; Parthasarathy, R.; Sawicki, G.; Lin, X-M.; Jaeger, H.; Lindquist, S.L. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc. Natl. Acad. Sci. USA 2003, 100, 4527–4532. [Google Scholar] [CrossRef] [PubMed]
- Domigan, L.J.; Healy, J.P.; Meade, S.J.; Blaikie, R.J.; Gerrard, J.A. Controlling the dimensions of amyloid fibrils: Toward homogenous components for bionanotechnology. Biopolymers 2011, 97, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Bolisetty, S.; Adamcik, J.; Heier, J.; Mezzenga, R. Amyloid directed synthesis of titanium dioxide nanowires and their applications in hybrid photovoltaic devices. Adv. Funct. Mater. 2012, 22, 3424–3428. [Google Scholar] [CrossRef]
- Rao, S.P.; Meade, S.J.; Healy, J.P.; Sutton, K.H.; Larsen, N.G.; Staiger, M.P.; Gerrard, J.A. Amyloid fibrils as functionalizable components of nanocomposite materials. Biotechnol. Prog. 2012, 28, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Bolisetty, S.; Mezzenga, R. Hybrid nanocomposites of gold single- crystal platelets and amyloid fibrils with tunable fluorescence, conductivity, and sensing properties. Adv. Mater. 2013, 25, 3694–3700. [Google Scholar] [CrossRef] [PubMed]
- Knowles, T.P.J.; Oppenheim, T.W.; Buell, A.K.; Chirgadze, D.Y.; Welland, M.E. Nanostructured films from hierarchical self-assembly of amyloidogenic proteins. Nat. Nanotechnol. 2010, 5, 204–207. [Google Scholar] [CrossRef] [PubMed]
- Ahn, M.; Kang, S.; Koo, H.J.; Lee, J.H.; Lee, Y.S.; Paik, S.R. Nanoporous protein matrix made of amyloid fibrils of β2-microglobulin. Biotechnol. Prog. 2010, 64, 1759–1764. [Google Scholar] [CrossRef] [PubMed]
- Bhak, G.; Lee, S.; Park, J.W.; Cho, S.; Paik, S.R. Amyloid hydrogel derived from curly protein fibrils of alpha-synuclein. Biomaterials 2010, 31, 5986–5995. [Google Scholar] [CrossRef] [PubMed]
- Nystrom, G.; Fernandez-Ronco, M.P.; Bolisetty, S.; Mazzotti, M.; Mezzenga, R. Amyloid templated gold aerogels. Adv. Mater. 2016, 28, 472. [Google Scholar] [CrossRef] [PubMed]
- Kaur, M.; Healy, J.; Vasudevamurthy, M.; Lassé, M.; Puskar, L.; Tobin, M.J.; Valery, C.; Gerrard, J.A.; Sasso, L. Stability and cytotoxicity of crystallin amyloid nanofibrils. Nanoscale 2014, 6, 13169–13178. [Google Scholar] [CrossRef] [PubMed]
- Bouchard, M.; Zurdo, J.; Nettleton, E.J.; Dobson, C.M.; Robinson, C.V. Formation of insulin amyloid fibrils followed by FTIR simultaneously with CD and electron microscopy. Protein Sci. 2000, 9, 1960–1967. [Google Scholar] [CrossRef] [PubMed]
- Loveday, S.M.; Su, J.; Rao, M.A.; Anema, S.G.; Singh, H. Effect of calcium on the morphology and functionality of whey protein nanofibrils. Biomacromolecules 2011, 12, 3780–3788. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.-H.; Wang, C.-S. Formation and characterization of amyloid-like fibrils from soy β- conglycinin and glycinin. J. Agric. Food Chem. 2010, 58, 11058–11066. [Google Scholar]
- Garvey, M.; Gras, S.L.; Meehan, S.; Meade, S.J.; Carver, J.A.; Gerrard, J.A. Protein nanofibres of defined morphology prepared from mixtures of crude crystallins. Int. J. Nanotechnol. 2009, 6, 258. [Google Scholar] [CrossRef]
- Healy, J.; Wong, K.; Sawyer, E.B.; Roux, C.; Domigan, L.; Gras, S.L.; Sunde, M.; Larsen, N.G.; Gerrard, J.; Vasudevamurthy, M. Polymorphism and higher order structures of protein nanofibers from crude mixtures of fish lens crystallins: Toward useful materials. Biopolymers 2012, 97, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Kang, S.; Lee, S.-G.; Jin, J.-H.; Park, J.W.; Park, S.M.; Jung, S.; Paik, S.R. Fibrillar superstructure formation of hemoglobin A and its conductive, photodynamic and photovoltaic effects. Acta Biomater. 2010, 6, 4689–4697. [Google Scholar] [CrossRef] [PubMed]
- Iram, A.; Alam, T.; Khan, J.M.; Khan, T.A.; Khan, R.H.; Naeem, A. Molten globule of hemoglobin proceeds into aggregates and advanced glycated end products. PLoS ONE 2013, 8, e72075. [Google Scholar] [CrossRef] [PubMed]
- Iram, A.; Naeem, A. Detection and analysis of protofibrils and fibrils of hemoglobin: Implications for the pathogenesis and cure of heme loss related maladies. Arch. Biochem. Biophys. 2013, 533, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Sunde, M.; Serpell, L.C.; Bartlam, M.; Fraser, P.E.; Pepys, M.B.; Blake, C.C. Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol. 1997, 273, 729–739. [Google Scholar] [CrossRef] [PubMed]
- Makin, O.S.; Serpell, L.C. Structures for amyloid fibrils. FEBS J. 2005, 272, 5950–5961. [Google Scholar] [CrossRef] [PubMed]
- Goldsbury, C.; Kistler, J.; Aebi, U.; Arvinte, T.; Cooper, G.J. Watching amyloid fibrils grow by time-lapse atomic force microscopy. J. Mol. Biol. 1999, 285, 33–39. [Google Scholar] [CrossRef] [PubMed]
- Serpell, L.C.; Sunde, M.; Benson, M.D.; Tennent, G.A.; Pepys, M.B.; Fraser, P.E. The protofilament substructure of amyloid fibrils. J. Mol. Biol. 2000, 300, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, J.L.; Nettleton, E.J.; Bouchard, M.; Robinson, C.V.; Dobson, C.M.; Saibil, H.R. The protofilament structure of insulin amyloid fibrils. Proc. Natl. Acad. Sci. USA 2002, 99, 9196–9201. [Google Scholar] [CrossRef] [PubMed]
- Adamcik, J.; Jung, J.M.; Flakowski, J.; De Los Rios, P.; Dietler, G.; Mezzenga, R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat. Nanotechnol. 2010, 5, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Holmes, T.C.; DiPersio, C.M.; Hynes, R.O.; Su, X.; Rich, A. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials 1995, 16, 1385–1393. [Google Scholar] [CrossRef]
- Reches, M. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science 2003, 300, 625–627. [Google Scholar] [CrossRef] [PubMed]
- Domigan, L.J. New Nanomaterials: Amyloid Fibrils from Waste Protein. Ph.D. Thesis, University of Canterbury, Christchurch, New Zealand, 2012. [Google Scholar]
- Jordens, S.; Adamcik, J.; Amar-Yuli, I.; Mezzenga, R. Disassembly and reassembly of amyloid fibrils in water-ethanol mixtures. Biomacromolecules 2011, 12, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Hirota-nakaoka, N.; Hasegawa, K.; Naiki, H.; Goto, Y. Dissolution of β2 -microglobulin amyloid fibrils by dimethylsulfoxide. J. Biochem. 2003, 134, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, B.; Pras, M. Preparative fractionation of amyloid proteins on a microgram scale by high-performance liquid chromatography and polyacrylamide gel electrophoresis. Clin. Chim. Acta. 1987, 163, 199–205. [Google Scholar] [CrossRef]
- Surmacz-Chwedoruk, W.; Malka, I.; Bożycki, Ł.; Nieznańska, H.; Dzwolak, W. On the heat stability of amyloid-based biological activity: Insights from thermal degradation of insulin fibrils. PLoS ONE 2014, 9, e86320. [Google Scholar] [CrossRef] [PubMed]
- Gras, S.L.; Tickler, A.K.; Squires, A.M.; Devlin, G.L.; Horton, M.A.; Dobson, C.M.; MacPhee, C.E. Functionalized amyloid fibrils for roles in cell adhesion. Biomaterials 2008, 29, 1553–1562. [Google Scholar] [CrossRef] [PubMed]
- Hackert, M.L.; Riggs, A.F. When size matters. Structure 2006, 14, 1094–1096. [Google Scholar] [CrossRef] [PubMed]
- Xue, W.-F.; Hellewell, A.L.; Gosal, W.S.; Homans, S.W.; Hewitt, E.W.; Radford, S.E. Fibril fragmentation enhances amyloid cytotoxicity. J. Biol. Chem. 2009, 284, 34272–34282. [Google Scholar] [CrossRef] [PubMed]
- Mankar, S.; Anoop, A.; Sen, S.; Maji, S.K. Nanomaterials: Amyloids reflect their brighter side. Nano Rev. 2011, 2, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schmittschmitt, J.P.; Scholtz, J.M. The role of protein stability, solubility, and net charge in amyloid fibril formation. Protein Sci. 2013, 12, 2374–2378. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.S. Protease-resistant human prion protein and ferritin are cotransported across Caco-2 epithelial cells: Implications for species barrier in prion uptake from the intestine. J. Neurosci. 2004, 24, 11280–11290. [Google Scholar] [CrossRef] [PubMed]
- Söderberg, L.; Dahlqvist, C.; Kakuyama, H.; Thyberg, J.; Ito, A.; Winblad, B.; Näslund, J.; Tjernberg, L.O. Collagenous Alzheimer amyloid plaque component assembles amyloid fibrils into protease resistant aggregates. FEBS J. 2005, 272, 2231–2236. [Google Scholar] [CrossRef] [PubMed]
- Langer, F.; Eisele, Y.S.; Fritschi, S.K.; Staufenbiel, M.; Walker, L.C.; Jucker, M. Soluble Aβ seeds are potent inducers of cerebral β-amyloid deposition. Neurosci. 2011, 31, 14488–14495. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.C.; Stöhr, J.; Bhardwaj, S.; Wille, H.; Oehler, A.; Dearmond, S.J.; Giles, K.; Prusiner, S.B. Protease-resistant prions selectively decrease Shadoo protein. PLoS Pathog. 2011, 7, e1002382. [Google Scholar] [CrossRef] [PubMed]
- Saido, T.; Leissring, M.A. Proteolytic degradation of amyloid β-protein. Cold Spring Harb. Perspect. Med. 2012, 2, a006379. [Google Scholar] [CrossRef] [PubMed]
- Lassé, M.; Ulluwishewa, D.; Healy, J.; Thompson, D.; Miller, A.; Roy, N.; Chitcholtan, K.; Gerrard, J.A. Evaluation of protease resistance and toxicity of amyloid- like food fibrils from whey, soy, kidney bean, and egg white. Food Chem. 2016, 192, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.Y. Scaling up the Production of Protein Nanofibers. MEng Thesis, University of Canterbury, New Zealand, 2011. [Google Scholar]
- Arias, M.; Quijano, J.C.; Haridas, V.; Gutterman, J.U.; Lemeshko, V.V. Red blood cell permeabilization by hypotonic treatments, saponin, and anticancer avicins. Acta Biomembr. 2010, 1798, 1189–1196. [Google Scholar] [CrossRef] [PubMed]
- Fens, M.H.A.M.; van Wijk, R.; Andringa, G.; van Rooijen, K.L.; Dijstelbloem, H.M.; Rasmussen, J.T.; de Vooght, K.M.K.; Schiffelers, R.M.; Gaillard, C.A.J.M.; van Solinge, W.W. A role for activated endothelial cells in red blood cell clearance: Implications for vasopathology. Haematologica 2012, 97, 500–508. [Google Scholar] [CrossRef] [PubMed]
- Keohane, E.M.; Smith, L.J.; Walenga, J.M. Rodak’s Hematology: Clinical Principles and Applications, 5th ed.; Elsevier Inc.: Amsterdam, Netharlands, 2016; pp. 372–373. [Google Scholar]
- Ascoli, F.; Rosaria, M.; Fanelli, R.; Antonini, E. Preparation and properties of apohemoglobin and reconstituted hemoglobins. Methods Enzymol. 1981, 76, 72–87. [Google Scholar] [PubMed]
- LeVine, H. Thioflavin T interaction with synthetic Alzheimer’s disease beta- amyloid peptides: Detection of amyloid aggregation in solution. Protein Sci. 1993, 2, 404–410. [Google Scholar] [CrossRef] [PubMed]
- Nečas, D.; Klapetek, P. Gwyddion: An open source software for SPM data analysis. Cent. Eur. J. Phys. 2012, 10, 181–188. [Google Scholar]
- Taylor, A.A.; Mendis, B.G. TEM characterzation of biological and inorganic nanocomposites. In Transmission Electron Microscopy Characterization of Nanomaterials, 1st ed.; Springer: Berlin, Germany, 2014; pp. 43–88. [Google Scholar]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.L.; Serpell, L.C. X-ray fiber diffraction studies of amyloid fibrils. Methods Mol. Biol. 2012, 849, 121–135. [Google Scholar] [PubMed]
- Hsieh, S.; Hsieh, C. Alignment of gold nanoparticles using insulin fibrils as a sacrificial biotemplate. Chem. Commun. 2010, 46, 7355–7357. [Google Scholar] [CrossRef] [PubMed]







© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jayawardena, N.; Kaur, M.; Nair, S.; Malmstrom, J.; Goldstone, D.; Negron, L.; Gerrard, J.A.; Domigan, L.J. Amyloid Fibrils from Hemoglobin. Biomolecules 2017, 7, 37. https://doi.org/10.3390/biom7020037
Jayawardena N, Kaur M, Nair S, Malmstrom J, Goldstone D, Negron L, Gerrard JA, Domigan LJ. Amyloid Fibrils from Hemoglobin. Biomolecules. 2017; 7(2):37. https://doi.org/10.3390/biom7020037
Chicago/Turabian StyleJayawardena, Nadishka, Manmeet Kaur, Smitha Nair, Jenny Malmstrom, David Goldstone, Leonardo Negron, Juliet A. Gerrard, and Laura J. Domigan. 2017. "Amyloid Fibrils from Hemoglobin" Biomolecules 7, no. 2: 37. https://doi.org/10.3390/biom7020037
APA StyleJayawardena, N., Kaur, M., Nair, S., Malmstrom, J., Goldstone, D., Negron, L., Gerrard, J. A., & Domigan, L. J. (2017). Amyloid Fibrils from Hemoglobin. Biomolecules, 7(2), 37. https://doi.org/10.3390/biom7020037