Cooperative Cellular Uptake and Activity of Octaarginine Antisense Peptide Nucleic Acid (PNA) Conjugates
Abstract
:1. Introduction
2. Materials and Methods
2.1. Peptide Nucleic Acid (PNA) Synthesis
2.2. Cell Culture
2.3. PNAs Antisense Activity and Toxicity
2.4. Reverse Transcriptase Polymerase Chain Reaction
2.5. Fluorescence and Confocal Microscopy
2.6. Flow Cytometry
2.7. Cellular Uptake by Fluorescence Reading in 96-Well Plates
2.8. Size of Self-Aggregated Particles
2.9. Characterization of PNA–Protein Interaction
3. Results
3.1. Cooperative PNA Activity
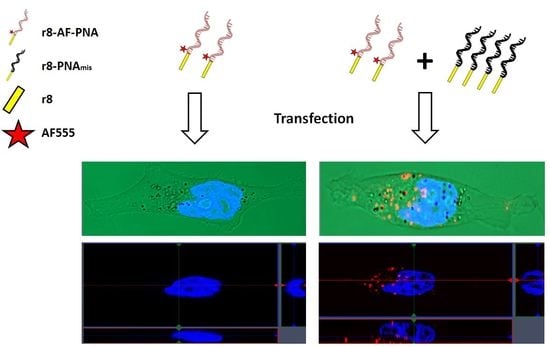
3.2. Cellular Uptake of PNA Using Fluorescence Microscopy
3.3. Cellular Uptake Quantification by Flow Cytometry
3.4. Enhancing Effect of Deca-r8-PNA on Non-Peptide PNA Derivatives
3.5. Nano-Aggregate/Particle Formation in the Presence of Serum
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Shiraishi, T.; Eysturskarth, J.; Nielsen, P.E. Modulation of MDM2 pre-mRNA splicing by 9-aminoacridine-PNA (peptide nucleic acid) conjugates targeting intron-exon junctions. BMC Cancer 2010, 10, 342. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Nielsen, P.E. Down-regulation of MDM2 and activation of p53 in human cancer cells by antisense 9-aminoacridine-PNA (peptide nucleic acid) conjugates. Nucleic Acids Res. 2004, 32, 4893–4902. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiraishi, T.; Hamzavi, R.; Nielsen, P.E. Subnanomolar antisense activity of phosphonate-peptide nucleic acid (PNA) conjugates delivered by cationic lipids to HeLa cells. Nucleic Acids Res. 2008, 36, 4424–4432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pooga, M.; Soomets, U.; Hällbrink, M.; Valkna, A.; Saar, K.; Rezaei, K.; Kahl, U.; Hao, J.X.; Xu, X.J.; Wiesenfeld-Hallin, Z.; et al. Cell penetrating PNA constructs regulate galanin receptor levels and modify pain transmission in vivo. Nat. Biotechnol. 1998, 16, 857–861. [Google Scholar] [CrossRef] [PubMed]
- Fraser, G.L.; Holmgren, J.; Clarke, P.B.; Wahlestedt, C. Antisense inhibition of δ-opioid receptor gene function in vivo by peptide nucleic acids. Mol. Pharmacol. 2000, 57, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Lu, Q.; Wood, M. Effective exon skipping and restoration of dystrophin expression by peptide nucleic acid antisense oligonucleotides in MDX mice. Mol. Ther. 2008, 16, 38–45. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.M.; Sahu, B.; Rapireddy, S.; Bahal, R.; Wheeler, S.E.; Procopio, E.M.; Kim, J.; Joyce, S.C.; Contrucci, S.; Wang, Y.; et al. Antitumor effects of EGFR antisense guanidine-based peptide nucleic acids in cancer models. ACS Chem. Biol. 2013, 8, 345–352. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, P.E.; Egholm, M. An introduction to peptide nucleic acid. Curr. Issues Mol. Biol. 1999, 1, 89–104. [Google Scholar]
- Shiraishi, T.; Bendifallah, N.; Nielsen, P.E. Cellular delivery of polyheteroaromate-peptide nucleic acid conjugates mediated by cationic lipids. Bioconjug. Chem. 2006, 17, 189–194. [Google Scholar] [CrossRef]
- Doyle, D.F.; Braasch, D.A.; Simmons, C.G.; Janowski, B.A.; Corey, D.R. Inhibition of gene expression inside cells by peptide nucleic acids: Effect of mRNA target sequence, mismatched bases, and PNA length. Biochemistry 2001, 40, 53–64. [Google Scholar] [CrossRef]
- Lv, H.; Zhang, S.; Wang, B.; Cui, S.; Yan, J. Toxicity of cationic lipids and cationic polymers in gene delivery. J. Control. Release 2006, 114, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Shiraishi, T.; Nielsen, P.E. Peptide nucleic acid (PNA) cell penetrating peptide (CPP) conjugates as carriers for cellular delivery of antisense oligomers. Artif. DNA PNA XNA 2011, 2, 90–99. [Google Scholar] [PubMed] [Green Version]
- Palm-Apergi, C.; Lonn, P.; Dowdy, S.F. Do cell-penetrating peptides actually “penetrate” cellular membranes? Mol. Ther. 2012, 20, 695–697. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, A.; Futaki, S.; Harashima, H. Delivery of Macromolecules Using Arginine-Rich Cell-Penetrating Peptides: Ways to Overcome Endosomal Entrapment. AAPS J. 2009, 11, 13–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margus, H.; Padari, K.; Pooga, M. Cell-penetrating peptides as versatile vehicles for oligonucleotide delivery. Mol. Ther. 2012, 20, 525–533. [Google Scholar] [CrossRef] [PubMed]
- Richard, J.P.; Melikov, K.; Vives, E.; Ramos, C.; Verbeure, B.; Gait, M.J.; Chernomordik, L.V.; Lebleu, B. Cell-penetrating peptides. A reevaluation of the mechanism of cellular uptake. J. Biol. Chem. 2003, 278, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Nakase, I.; Niwa, M.; Takeuchi, T.; Sonomura, K.; Kawabata, N.; Koike, Y.; Takehashi, M.; Tanaka, S.; Ueda, K.; Simpson, J.C.; et al. Cellular uptake of arginine-rich peptides: Roles for macropinocytosis and actin rearrangement. Mol. Ther. 2004, 10, 1011–1022. [Google Scholar] [CrossRef]
- Järver, P.; Langel, K.; El-Andaloussi, S.; Langel, Ü. Applications of cell-penetrating peptides in regulation of gene expression. Biochem. Soc. Trans. 2007, 35, 770–774. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, A.; Dowdy, S.F. Protein transduction domain delivery of therapeutic macromolecules. Curr. Opin. Biotechnol. 2011, 22, 888–893. [Google Scholar] [CrossRef]
- Maiolo, J.R.; Ferrer, M.; Ottinger, E.A. Effects of cargo molecules on the cellular uptake of arginine-rich cell-penetrating peptides. Biochim. Biophys. Acta 2005, 1712, 161–172. [Google Scholar] [CrossRef] [Green Version]
- Shiraishi, T.; Pankratova, S.; Nielsen, P.E. Calcium ions effectively enhance the effect of antisense peptide nucleic acids conjugated to cationic tat and oligoarginine peptides. Chem. Biol. 2005, 12, 923–929. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.J.; Ivanova, G.D.; Verbeure, B.; Williams, D.; Arzumanov, A.A.; Abes, S.; Lebleu, B.; Gait, M.J. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Res. 2005, 33, 6837–6849. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarani, R.; Shiraishi, T.; Nielsen, P.E. Effective photo-enhancement of cellular activity of fluorophore-octaarginine antisense PNA conjugates correlates with singlet oxygen formation, endosomal escape and chromophore lipophilicity. Sci. Rep. 2018, 8, 638. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, S.M.; Raines, R.T. Pathway for Polyarginine Entry into Mammalian Cells. Biochemistry 2004, 43, 2438–2444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppelhus, U.; Shiraishi, T.; Zachar, V.; Pankratova, S.; Nielsen, P.E. Improved cellular activity of antisense peptide nucleic acids by conjugation to a cationic peptide-lipid (CatLip) domain. Bioconjug. Chem. 2008, 19, 1526–1534. [Google Scholar] [CrossRef]
- Christensen, L.; Fitzpatrick, R.; Gildea, B.; Petersen, K.H.; Hansen, H.F.; Koch, T.; Egholm, M.; Buchardt, O.; Nielsen, P.E.; Coull, J.; et al. Solid-phase synthesis of peptide nucleic acids. J. Pept. Sci. 1995, 1, 175–183. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Kang, S.H.; Cho, M.J.; Kole, R. Up-regulation of luciferase gene expression with antisense oligonucleotides: Implications and applications in functional assay development. Biochemistry 1998, 37, 6235–6239. [Google Scholar] [CrossRef]
- Ezzat, K.; ELAndaloussi, S.; Zaghloul, E.M.; Lehto, T.; Lindberg, S.; Moreno, P.M.; Viola, J.R.; Magdy, T.; Abdo, R.; Guterstam, P.; et al. PepFect 14, a novel cell-penetrating peptide for oligonucleotide delivery in solution and as solid formulation. Nucleic Acids Res. 2011, 39, 5284–5298. [Google Scholar] [CrossRef]
- Valero, J.; Shiraishi, T.; de Mendoza, J.; Nielsen, P.E. Cellular Antisense Activity of PNA-Oligo(bicycloguanidinium) Conjugates Forming Self-Assembled Nanoaggregates. Chembiochem 2015, 16, 1593–1600. [Google Scholar] [CrossRef]
- Ezzat, K.; Aoki, Y.; Koo, T.; McClorey, G.; Benner, L.; Coenen-Stass, A.; O’Donovan, L.; Lehto, T.; Garcia-Guerra, A.; Nordin, J.; et al. Self-Assembly into Nanoparticles Is Essential for Receptor Mediated Uptake of Therapeutic Antisense Oligonucleotides. Nano Lett. 2015, 15, 4364–4373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guidotti, G.; Brambilla, L.; Rossi, D. Cell-Penetrating Peptides: From Basic Research to Clinics. Trends Pharmacol. Sci. 2017, 38, 406–424. [Google Scholar] [CrossRef] [PubMed]
- Behzadi, S.; Serpooshan, V.; Tao, W.; Hamaly, M.A.; Alkawareek, M.Y.; Dreaden, E.C.; Brown, D.; Alkilany, A.M.; Farokhzad, O.C.; Mahmoudi, M. Cellular uptake of nanoparticles: Journey inside the cell. Chem. Soc. Rev. 2017, 46, 4218–4244. [Google Scholar] [CrossRef] [PubMed]
- Cardarelli, F.; Digiacomo, L.; Marchini, C.; Amici, A.; Salomone, F.; Fiume, G.; Rossetta, A.; Gratton, E.; Pozzi, D.; Caracciolo, G. The intracellular trafficking mechanism of Lipofectamine-based transfection reagents and its implication for gene delivery. Sci. Rep. 2016, 6, 25879. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; DeRidder, L.; Elmore, B.; Dong, H. Self-assembly of Filamentous Cell Penetrating Peptides for Gene Delivery. Methods Mol. Biol. 2018, 1777, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Weinberger, A.; Walter, V.; MacEwan, S.R.; Schmatko, T.; Muller, P.; Schroder, A.P.; Chilkoti, A.; Marques, C.M. Cargo self-assembly rescues affinity of cell-penetrating peptides to lipid membranes. Sci. Rep. 2017, 7, 43963. [Google Scholar] [CrossRef] [Green Version]
- Emi, N.; Kidoaki, S.; Yoshikawa, K.; Saito, H. Gene transfer mediated by polyarginine requires a formation of big carrier-complex of DNA aggregate. Biochem. Biophys. Res. Commun. 1997, 231, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Kushnaryov, V.M.; MacDonald, H.S.; Sedmak, J.J.; Grossberg, S.E. Murine interferon-β receptor-mediated endocytosis and nuclear membrane binding. Proc. Natl. Acad. Sci. USA 1985, 82, 3281–3285. [Google Scholar] [CrossRef]
- Sorkin, A. Cargo recognition during clathrin-mediated endocytosis: A team effort. Curr. Opin. Cell Biol. 2004, 16, 392–399. [Google Scholar] [CrossRef]
- Traub, L.M.; Bonifacino, J.S. Cargo recognition in clathrin-mediated endocytosis. Cold Spring Harb. Perspect. Biol. 2013, 5, a016790. [Google Scholar] [CrossRef]
- Yue, T.; Zhang, X. Cooperative Effect in Receptor-Mediated Endocytosis of Multiple Nanoparticles. ACS Nano 2012, 6, 3196–3205. [Google Scholar] [CrossRef] [PubMed]








| PNA | Name | Sequence |
|---|---|---|
| 2389 | PNA | H-CCT CTT ACC TCA GTT ACA-NH2 |
| 5006 | r8-PNA | H-(D-Arg)8-CCT CTT ACC TCA GTT ACA-NH2 |
| 2784 | Deca-r8-PNA | H-(D-Arg)8-Lys(Decanoyl)- CCT CTT ACC TCA GTT ACA-NH2 |
| 3388 | Deca-r8-PNAmm | H-(D-Arg)8-Lys(Decanoyl)- CCT CTG ACC TCA TTT ACA-NH2 |
| 5102 | AF-PNA | Ac-Cys(AF555)-CCT CTT ACC TCA GTT ACA-NH2 |
| 5103 | Deca-r8-AF-PNA | H-(D-Arg)8-Lys(Decanoyl)-Cys(AF555)-CCT CTTT ACC TCA GTT ACA-NH2 |
| 5104 | r8-AF-PNA | H-(D-Arg)8-Cys(AF555)-CCT CTTT ACC TCA GTT ACA-NH2 |
| 5224 | r8-PNAmm | H-(D-Arg)8-CCT CTG ACC TCA TTT ACA-NH2 |
| 2964 | Cholate-PNA | Cholate-CCT CTT ACC TCA GTT ACA-NH2 |
| 5009 | Deca-r8-AF-PNA2 | Decanoyl-(D-Arg)8-Cys(AF555)-TCC AGA TGC CTT GGG-NH2 |
| 5012 | Deca-r8-PNA2 | Decanoyl-(D-Arg)8-Cys-TCC AGA TGC CTT GGG-NH2 |
| Pep44 | Deca-r8 | Decanoyl-(D-Arg)8-Gly-NH2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ghavami, M.; Shiraishi, T.; Nielsen, P.E. Cooperative Cellular Uptake and Activity of Octaarginine Antisense Peptide Nucleic Acid (PNA) Conjugates. Biomolecules 2019, 9, 554. https://doi.org/10.3390/biom9100554
Ghavami M, Shiraishi T, Nielsen PE. Cooperative Cellular Uptake and Activity of Octaarginine Antisense Peptide Nucleic Acid (PNA) Conjugates. Biomolecules. 2019; 9(10):554. https://doi.org/10.3390/biom9100554
Chicago/Turabian StyleGhavami, Mahdi, Takehiko Shiraishi, and Peter E. Nielsen. 2019. "Cooperative Cellular Uptake and Activity of Octaarginine Antisense Peptide Nucleic Acid (PNA) Conjugates" Biomolecules 9, no. 10: 554. https://doi.org/10.3390/biom9100554
APA StyleGhavami, M., Shiraishi, T., & Nielsen, P. E. (2019). Cooperative Cellular Uptake and Activity of Octaarginine Antisense Peptide Nucleic Acid (PNA) Conjugates. Biomolecules, 9(10), 554. https://doi.org/10.3390/biom9100554