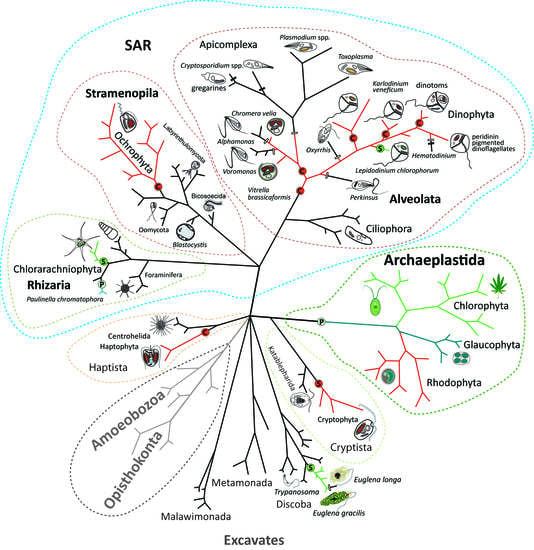
Endosymbiotic Evolution of Algae, Secondary Heterotrophy and Parasitism
Abstract
:1. Introduction
2. Primary Endosymbioses
3. Complex Endosymbioses
4. Secondary Heterotrophy and Parasitism in Algae
5. Conclusions
Funding
Acknowledgments
Conflicts of Interest
References
- Shih, P.M. Cyanobacterial evolution: Fresh insight into ancient questions. Curr. Biol. 2015, 25, R193. [Google Scholar] [CrossRef] [PubMed]
- Shih, P.M.; Matzke, J. Primary endosymbiosis events date to the later Proterozoic with cross-calibrated phylogenetic dating of duplicated ATPase proteins. Proc. Natl. Acad. Sci USA 2013, 110, 12355–12360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mereschkowksy, C. Ober Natur und Ursprung der Chromatophoren im Pflanzenreiche. Biol. Zentralbl. 1905, 25, 593–604. [Google Scholar]
- Martin, W.; Kowallik, K.V. Annotated English translation of of Mereschkowsky’s 1905 paper “Über natur und Ursprung der Chromatophoren im Pflanzenreiche”. Eur. J. Phycol. 1999, 34, 287–295. [Google Scholar] [CrossRef]
- Pallen, M.J. Time to recognize that mitochondria are bacteria? Trends Microbiol. 2011, 19, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Oborník, M. In the beginning was the word: How terminology drives our understanding of endosymbiotic organelles. Microbial Cell 2019, 6, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T.; Lee, J.J. Protozoa as hosts for endosymbioses and the conversion of symbionts into organelles. J. Protozool. 1985, 32, 376–379. [Google Scholar] [CrossRef]
- Gould, S.B.; Waller, R.F.; McFadden, G.I. Plastid evolution. Ann. Rev. Plant. Biol. 2008, 59, 491–517. [Google Scholar] [CrossRef]
- Keeling, P.J. The number, speed, and impact of plastid endosymbioses in eukaryotic evolution. Ann. Rev. Plant. Biol. 2013, 64, 583–607. [Google Scholar] [CrossRef]
- Archibald, J.M. Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 2015, 25, R911–R921. [Google Scholar] [CrossRef]
- Gruber, A. What`s a name? Why organelles of endosymbiotic origin are implicitly called by their eukaryotic species name and how they can be distinguished from endosymbionts. Microbial Cell 2019, 6, 123–133. [Google Scholar] [CrossRef]
- Keeling, P.J. Diversity and evolutionary history of plastids and their hosts. Am. J. Bot. 2004, 91, 1481–1493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackson, C.; Clayden, S.; Reyes-Prieto, A. The Glaucophyta: The blue-green plants in a nutshell. Acta Soc. Bot. Pol. 2015, 84, 149–165. [Google Scholar] [CrossRef]
- Maréchal, E. (Ed.) Plastids: Methods and protocols. Methods in molecular biology. In Primary Endosymbiosis: Emergence of Primary Chloroplasts and Chromatophore Two Independent Events; Humana Press: New York, NY, USA, 2018; Volume 1829. [Google Scholar]
- Larkum, A.W.D.; Kühl, M. Chlorophyll d: The puzzle resolved. Trends Plant. Sci. 2005, 10, P355–P357. [Google Scholar] [CrossRef] [PubMed]
- Martin, W.; Herrmann, R.G. Gene transfer from organelles to the nucleus: How much, what happens, and why? Plant. Phys. 1998, 118, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Oborník, M.; Green, B. Mosaic origin of the heme biosynthesis pathway in photosynthetic eukaryotes. Mol. Biol. Evol. 2005, 22, 2343–2353. [Google Scholar] [CrossRef] [PubMed]
- Cihlář, J.; Füssy, Z.; Oborník, M. Evolution of tetrapyrrole pathway in eukaryotic phototrophs. Adv. Bot. Res. 2019, 90, 273–309. [Google Scholar]
- Cihlář, J.; Füssy, Z.; Horák, A.; Oborník, M. Evolution of the tetrapyrrole biosyntheses pathway in secondary algae: Conservation, redundancy, and replacement. PLoS ONE 2016, 11, e0166338. [Google Scholar] [CrossRef]
- Stiller, J.W.; Reel, D.C.; Johnson, J.C. A single origin of plastids revisited: Convergent evolution in organellar genome content. J. Phycol. 2003, 39, 95–105. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Principles of protein and lipid targeting in secondary symbiogenesis: Euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 1999, 46, 347–366. [Google Scholar] [CrossRef]
- Marin, B.; Nowack, E.C.M.; Melkonian, M. A plastid in the making: Evidence for a second primary endosymbiosis. Protist 2005, 156, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.S.; Reyes-Prieto, A.; Melkonian, M.; Bhattacharya, D. Minimal plastid genome evolution in the Paulinella endosymbiont. Curr. Biol. 2006, 16, R670–R672. [Google Scholar] [CrossRef] [PubMed]
- Delaye, L.; Valadez-Cano, C.; Pérez-Zamorano, B. How really ancient is Paulinella chromatophora? PLoS Curr. Tree of Life 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Delwiche, C.F.; Palmer, J.D. The Origin Plastids and Their Spread via Secondary Symbiosis Plant Systematics and Evolution; Springer: Berlin, Germany, 1997; pp. 53–86. (Suppl. 11). [Google Scholar]
- Delwiche, C.F. Tracing the thread of plastid diversity through the tapestry of life. Am. Nat. 1999, 154, S164–S177. [Google Scholar] [CrossRef] [PubMed]
- Falkowski, P.G.; Katz, M.E.; Knoll, A.H.; Quigg, A.; Raven, J.A.; Schofield, O.; Taylor, F.J. The evolution of modern phytoplankton. Science 2004, 305, 354–360. [Google Scholar] [CrossRef] [PubMed]
- Oborník, M. The birth of red complex plastids: One, three, or four times? Trends Parasitol. 2018, 34, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Curtis, B.; Tanifuji, G.; Burki, F.; Gruber, A.; Irimia, M.; Maruyama, S.; Arias, M.; Ball, S.; Gile, G.; Hirakawa, Y.; et al. Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 2012, 492, 59–65. [Google Scholar] [CrossRef]
- Vanclová, A.M.G.; Hadariová, L.; Hrdá, Š.; Hampl, V. Secondary plastids of euglenophytes. Adv. Bot. Res. 2017, 84, 321–358. [Google Scholar]
- Matsumoto, T.; Shinozaki, F.; Chikuni, T.; Yabuki, A.; Takishita, K.; Kawachi, M.; Nakayama, T.; Inouye, I.; Hashimoto, T.; Inagaki, Y. Green-colored plastids in the dinoflagellate genus Lepidodinium are of core chlorophyte origin. Protist 2011, 162, 268–276. [Google Scholar] [CrossRef]
- Waller, R.F.; Kořený, L. Plastid complexity on dinogflagellates: A picture of gains, losses, replacements and revisions. Adv. Bot. Res. 2017, 84, 105–143. [Google Scholar]
- Larkum, A.W.D.; Lockhart, P.J.; Howe, C.J. Shopping for plastids. Trends Plant. Sci. 2007, 12, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Cavalier-Smith, T. Kingdom Chromista and its eight phyla: A new synthesis emphasising periplastid protein targeting, cyto-skeletal and periplastid evolution, and ancient divergences. Protoplasma 2018, 255, 297–357. [Google Scholar] [CrossRef] [PubMed]
- Bodył, A.; Stiller, J.W.; Mackiewicz, P. Chromalveolate plastids: Direct descents or multiple endosymbioses? Trends Ecol. Evol. 2009, 24, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Stiller, J.W.; Schreiber, J.; Yue, J.; Guo, H.; Ding, Q.; Huang, J. The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Com. 2014, 5, 5764. [Google Scholar] [CrossRef] [PubMed]
- Petersen, J.; Ludewig, A.K.; Michael, V.; Bunk, B.; Jarek, M.; Baurain, D.; Brinkmann, H. Chromera velia, endosymbioses and the rhodoplex hypothesis—Plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages). Genome Biol. Evol. 2014, 6, 666–684. [Google Scholar] [CrossRef] [PubMed]
- Dorrell, D.R.; Bowler, C. Secondary plastids of stramenopiles. Adv. Bot. Res. 2017, 84, 57–103. [Google Scholar]
- Moore, C.E.; Archibald, J.M. Nucleomorph genomes. Ann. Rev. Genet. 2009, 43, 251–264. [Google Scholar] [CrossRef]
- Oborník, M.; Modrý, D.; Lukeš, M.; Černotíková-Stříbrná, E.; Cihlář, J.; Tesařová, M.; Kotabová, E.; Vancová, E.; Prášil, O.; Lukeš, J. Morphology, ultrastructure and life cycle of Vitrella brassicaformis n. sp., n. gen., a novel chromerid from the Great Barrier Reef. Protist 2012, 163, 306–323. [Google Scholar] [CrossRef]
- Ševčíková, T.; Horák, A.; Klimeš, V.; Zbránková, V.; Demir-Hilton, E.; Sudek, S.; Jenkins, J.; Schmutz, J.; Přibyl, P.; Fousek, J.; et al. Updating algal evolutionary relationships through plastid genome sequencing: Did alveolate plastids emerge through endosymbiosis of an ochrophyte? Sci. Rep. 2015, 5, 10134. [Google Scholar] [CrossRef] [PubMed]
- Sobotka, R.; Esson, H.J.; Koník, P.; Trsková, E.; Moravcová, L.; Horák, A.; Dufková, P.; Oborník, M. Extensive gain and loss of photosystem I subunits in chromerid algae, photosynthetic relatives of apicomplexans. Sci. Rep. 2017, 7, 13214. [Google Scholar] [CrossRef] [Green Version]
- Oborník, M.; Janouškovec, J.; Chrudimský, T.; Lukeš, J. Evolution of the apicoplast and its host: From heterotrophy to autotrophy and back again. Int. J. Parasitol. 2009, 39, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Pospíšil, P. Molecular mechanisms of production and scavenging of reactive oxygen species by photosystem II. Biochim. Biophys. Acta 2012, 1817, 218–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koblížek, M.; Zeng, Y.; Horák, A.; Oborník, M. Regressive evolution of photosynthesis in the Roseobacter clade. Adv. Bot. Res. 2013, 66, 385–405. [Google Scholar]
- Hadariová, L.; Vesteg, M.; Hampl, V.; Krajčovič, J. Reductive evolution of chloroplasts in non-photosynthetic plants, algae and protists. Curr. Genet. 2018, 64, 365–387. [Google Scholar] [CrossRef] [PubMed]
- Votýpka, J.; Nodrý, D.; Oborník, M.; Šlapeta, J.; Lukeš, J. Apicomplexa. In Handbook of the Protists; Archibald, J.M., Simpson, G.B., Slamovits, C.H., Eds.; Springer: Berlin, Germany, 2017; pp. 567–624. [Google Scholar]
- Těšitel, J. Functional biology of parasitic plants: A review. Plant. Ecol. Evol. 2017, 149, 5–20. [Google Scholar] [CrossRef]
- De la Cruz, V.F.; Gittleson, S.M. The genus Polytomella: A review of classification, morphology, life cycle, metabolism, and motility. Arch. Protistenkunde 1981, 124, 1–28. [Google Scholar] [CrossRef]
- Boucias, D.G.; Becnel, J.J.; White, S.E.; Bott, M. In vivo and in vitro development of the protist Helicosporidium sp. J. Eukaryot. Microbiol. 2001, 48, 460–470. [Google Scholar] [CrossRef] [PubMed]
- De Koning, A.P.; Keeling, P.J. The complete plastid genome sequence of the parasitic green alga Helicosporidium sp. is highly reduced and structured. BMC Biol. 2006, 4, 12. [Google Scholar] [CrossRef] [PubMed]
- Blouin, N.A.; Lane, C.E. Red algae provide fertile ground for exploring parasite evolution. Persp. Phycol. 2016, 2016. 3, 11–19. [Google Scholar] [CrossRef]
- Marin, B.; Palm, A.; Klingberg, M.; Melkonian, M. Phylogeny and taxonomic revision of plastid-containing euglenophytes based on SSU rDNA sequence comparisons and synapomorhic signatures in the SSU rRNA secondary structure. Protist 2003, 154, 99–145. [Google Scholar] [CrossRef]
- Lowe, C.D.; Keeling, P.J.; Martin, L.E.; Slamovits, C.H.; Watts, P.C.; Montagnes, D.J.S. Who is Oxyrrhis marina? Morphological and phylogenetic studies on an unusual dinoflagellate. J. Plankton Res. 2011, 33, 555–567. [Google Scholar] [CrossRef]
- Füssy, Z.; Oborník, M. Chromerids and their plastids. Adv. Bot. Res. 2017, 84, 187–218. [Google Scholar]
- Füssy, Z.; Oborník, M. Plastids: Methods and protocols methods in molecular biology. In Complex Endosymbioses I: From Primary to Complex Plastids, Multiple Independent Events; Maréchal, E., Ed.; Humana Press: New York, NY, USA, 2018; Volume 1829, pp. 17–35. [Google Scholar]
- Sato, S. The apicomplexan plastid and its evolution. Cell. Mol. Life Sci. 2011, 68, 1285–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toso, M.A.; Omoto, C.K. Gregarina niphandrodes may lack both a plastid genome and organelle. J. Euk. Microbiol. 2007, 54, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Gornik, S.G.; Febrimarsa, A.M.C.; MacRae, J.I.; Ramprasad, A.; Rchiad, Z.; McConville, M.J.; Bacic, A.; McFadden, G.I.; Pain, A.; Waller, R.F. Endosymbiosis undone by stepwise elimination of the plastid in a parasitic dinoflagellate. Proc. Natl. Acad. Sci. USA 2015, 112, 5767–5772. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burki, F. The convoluted evolution of eukaryotes with complex plastids. Adv. Bot. Res. 2017, 84, 1–30. [Google Scholar]
- Stoecker, D.K.; Hansen, P.J.; Caron, D.A.; Mitra, A. Mixotrophy in the marine plankton. Ann. Rev. Mar. Sci. 2016, 9, 311–335. [Google Scholar] [CrossRef]
- Rezic, T.; Filipović, J.; Santec, B. Photo-mixotrophic cultivation of algae Euglena gracilis for lipid production. Agr. Consp. Sci. 2013, 2013 78, 65–69. [Google Scholar]
- Heredia-Arroyo, T.; Wei, W.; Hu, B. Mixotrophic cultivation of Chlorella vulgaris and its potential application for the oil accumulation from non-sugar materials. Biomass Bioenerg. 2011, 35, 2245–2253. [Google Scholar] [CrossRef]
- Villanova, V.; Fortunato, A.E.; Singh, D.; Bo, D.D.; Conte, M.; Obata, T.; Jouhet, J.; Fernie, A.R.; Maréchal, E.; Falciatore, A.; et al. Investigating mixotrophic metabolism in the model diatom Phaeodactylum tricornutum. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160404. [Google Scholar] [CrossRef]
- Jeong, J.H.; Yoo, Y.D.; Kim, J.S.; Seong, K.A.; Kang, N.S.; Kim, T.H. Growth, feeding and ecological roles of the mixotrophic and heterotrophic dinoflagellates in marine planktonic food webs. Ocean Sci. J. 2010, 45, 65–91. [Google Scholar] [CrossRef] [Green Version]
- McFadden, G.I.; Reith, M.E.; Munholland, J.; Lang-Unnasch, N. Plastid in human parasites. Nature 1996, 381, 482. [Google Scholar] [CrossRef] [PubMed]
- McFadden, G.I.; Waller, R.F. Plastids in parasites of humans. Bioessays 1997, 19, 1033–1040. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.B.; Oborník, M.; Janouškovec, J.; Chrudimský, T.; Vancová, M.; Green, D.; Wright, S.; Davies, N.; Bolch, C.; Heimann, K.; et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature 2008, 451, 959–963. [Google Scholar] [CrossRef] [PubMed]
- Woo, Y.; Ansari, H.; Otto, T.D.; Klinger, C.; Kolísko, M.; Saxena, A.; Shanmugam, D.; Tayyrov, A.; Veluchamy, A.; Ali, S.; et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. elife 2015, 4, e06974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janouškovec, J.; Tikhonenkov, D.V.; Burki, F.; Howe, A.T.; Kolísko, M.; Mylnikov, A.P.; Keeling, P.J. Factors mediating plastid dependency and the origins of parasitism in apicomplexans and their close relatives. Proc. Natl. Acad. Sci. USA 2015, 112, 10200–10207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kořený, L.; Sobotka, R.; Janouškovec, J.; Keeling, P.J.; Oborník, M. Tetrapyrrole synthesis of photosynthetic chromerids is likely homologous to the unusual pathway of apicomplexan parasites. Plant. Cell 2011, 23, 3454–3462. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, N.; McFadden, G.I. The mother of all parasites. Futur. Microbiol. 2008, 3, 391–395. [Google Scholar] [CrossRef] [Green Version]
- Janouškovec, J.; Horák, A.; Barott, K.L.; Rohwer, F.L.; Keeling, P.J. Global analysis of plastid diversity reveals apicomplexan-related lineages in coral reefs. Curr. Biol. 2012, 22, R518–R519. [Google Scholar] [CrossRef] [Green Version]
- Janouškovec, J.; Horák, A.; Barott, K.L.; Rohwer, F.L.; Keeling, P.J. Environmental distribution of coral-associated relatives of apicomplexan parasites. ISME J. 2013, 7, 444–447. [Google Scholar] [CrossRef]
- Cumbo, V.R.; Baird, A.H.; Moore, R.B.; Negri, A.P.; Neilan, B.A.; Salih, A.; van Oppen, M.J.; Marquis, C.P. Chromera velia is endosymbiotic in larvae of the reef corals Acropora digitifera and A. tenuis. Protist 2013, 164, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, A.R.; Cumbo, V.R.; Harii, A.; Shizato, C.; Chan, C.X.; Ragan, M.A.; Satoh, N.; Ball, E.E.; Miller, D.J. Deciphering the nature of the coral-Chromera association. ISME J. 2018, 12, 776–790. [Google Scholar] [CrossRef] [PubMed]
- Skovgaard, A.; Karpov, S.A.; Guillou, L. The parasitic dinoflagellates Blastodinium spp. Inhabiting the gut of marine, plaktonic copepods: Morphology, ecology, and unrecognized species diversity. Front. Microbiol. 2012, 3, 305. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; McCutcheon, J.P. Endosymbiosis: The feeling is not mutual. J. Theor. Biol. 2017, 434, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Del Campo, J.; Mathur, V.; Vermeij, M.J.A.; Keeling, P.J. A widespread coral-infecting apicomplexan with chlorophyll biosynthesis genes. Nature 2019, 568, 103–107. [Google Scholar] [CrossRef] [PubMed]

© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oborník, M. Endosymbiotic Evolution of Algae, Secondary Heterotrophy and Parasitism. Biomolecules 2019, 9, 266. https://doi.org/10.3390/biom9070266
Oborník M. Endosymbiotic Evolution of Algae, Secondary Heterotrophy and Parasitism. Biomolecules. 2019; 9(7):266. https://doi.org/10.3390/biom9070266
Chicago/Turabian StyleOborník, Miroslav. 2019. "Endosymbiotic Evolution of Algae, Secondary Heterotrophy and Parasitism" Biomolecules 9, no. 7: 266. https://doi.org/10.3390/biom9070266
APA StyleOborník, M. (2019). Endosymbiotic Evolution of Algae, Secondary Heterotrophy and Parasitism. Biomolecules, 9(7), 266. https://doi.org/10.3390/biom9070266