The Relationship between AGAMOUS and Cytokinin Signaling in the Establishment of Carpeloid Features
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Hormone Treatments
2.3. Scanning Electron Microscopy
2.4. qRT-PCR Analysis
3. Results and Discussion
3.1. Exogenous Cytokinin Induces Carpeloid Features in an AG-Dependent Manner
3.2. Cytokinin Induces AG Targets Involved in Gynoecium Development
3.3. Different Effects of Type-B ARR Proteins on Carpel Promoting Genes
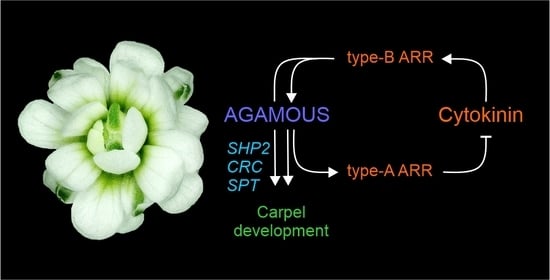
3.4. Model of Regulatory Network between AG and Cytokinin
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gaillochet, C.; Lohmann, J.U. The never-ending story: From pluripotency to plant developmental plasticity. Development 2015, 142, 2237–2249. [Google Scholar] [CrossRef] [Green Version]
- Smyth, D.R.; Bowman, J.L.; Meyerowitz, E.M. Early flower development in Arab. Plant Cell 1990, 2, 755–767. [Google Scholar] [PubMed] [Green Version]
- Alvarez-Buylla, E.R.; Benítez, M.; Corvera-Poiré, A.; Chaos Cador, A.; de Folter, S.; De Buen Gamboa, A.; Garay-Arroyo, A.; García-Ponce, B.; Jaimes-Miranda, F.; Pérez-Ruiz, R.V.; et al. Flower Development. Arab. Book 2010, 8, 1–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. The ABC model of flower development: Then and now. Development 2012, 4098, 4095–4098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denay, G.; Chahtane, H.; Tichtinsky, G.; Parcy, F. A flower is born: An update on Arabidopsis floral meristem formation. Curr. Opin. Plant Biol. 2017, 35, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Wils, C.R.; Kaufmann, K. Gene-regulatory networks controlling inflorescence and flower development in Arabidopsis thaliana. Biochim. Biophys. Acta-Gene Regul. Mech. 2017, 1860, 95–105. [Google Scholar] [CrossRef]
- Thomson, B.; Wellmer, F. Molecular regulation of flower development. Curr. Topics Dev. Biol. 2019, 131, 185–210. [Google Scholar]
- Zúñiga-Mayo, V.M.; Gómez-Felipe, A.; Herrera-Ubaldo, H.; de Folter, S. Gynoecium development: Networks in Arabidopsis and beyond. J. Exp. Bot. 2019, 70, 1447–1460. [Google Scholar] [CrossRef]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genes directing flower development in Arabidopsis. Plant Cell 1989, 1, 37–52. [Google Scholar]
- Yanofsky, M.F.; Ma, H.; Bowman, J.L.; Drews, G.N.; Feldmann, K.A.; Meyerowitz, E.M. The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 1990, 346, 35–39. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Dreni, L.; Kater, M.M. MADS reloaded: Evolution of the AGAMOUS subfamily genes. New Phytol. 2014, 201, 717–732. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [PubMed]
- Mizukami, Y.; Ma, H. Ectopic expression of the floral homeotic gene AGAMOUS in transgenic Arabidopsis plants alters floral organ identity. Cell 1992, 71, 119–131. [Google Scholar] [CrossRef]
- Bowman, J.L.; Drews, G.N.; Meyerowitz, E.M. Expression of the Arabidopsis floral homeotic gene AGAMOUS is restricted to specific cell types late in flower development. Plant Cell 1991, 3, 749–758. [Google Scholar]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar] [PubMed]
- Bowman, J.L.; Smyth, D.R. CRABS CLAW, a gene that regulates carpel and nectary development in Arabidopsis, encodes a novel protein with zinc finger and helix-loop-helix domains. Development 1999, 126, 2387–2396. [Google Scholar]
- Heisler, M.G.B.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar]
- Pinyopich, A.; Ditta, G.S.; Savidge, B.; Liljegren, S.J.; Baumann, E.; Wisman, E.; Yanofsky, M.F. Assessing the redundancy of MADS-box genes during carpel and ovule development. Nature 2003, 424, 85–88. [Google Scholar] [CrossRef]
- Favaro, R.; Pinyopich, A.; Battaglia, R.; Kooiker, M.; Borghi, L.; Ditta, G.; Yanofsky, M.F.; Kater, M.M.; Colombo, L. MADS-Box protein complexes control carpel and ovule developement in Arabidopsis. Plant Cell 2003, 15, 2603–2611. [Google Scholar] [CrossRef] [Green Version]
- Groszmann, M.; Paicu, T.; Alvarez, J.P.; Swain, S.M.; Smyth, D.R. SPATULA and ALCATRAZ, are partially redundant, functionally diverging bHLH genes required for Arabidopsis gynoecium and fruit development. Plant J. 2011, 68, 816–829. [Google Scholar] [CrossRef]
- Gross, T.; Broholm, S.; Becker, A. CRABS CLAW acts as a bifunctional transcription factor in flower development. Front. Plant Sci. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Ferrándiz, C.; Fourquin, C.; Prunet, N.; Scutt, C.P.; Sundberg, E.; Trehin, C.; Vialette-Guiraud, A.C.M. Carpel development. Adv. Bot. Res. 2010, 55, 1–73. [Google Scholar] [CrossRef]
- Gómez-Mena, C.; de Folter, S.; Costa, M.M.R.; Angenent, G.C.; Sablowski, R. Transcriptional program controlled by the floral homeotic gene AGAMOUS during early organogenesis. Development 2005, 132, 429–438. [Google Scholar] [CrossRef] [Green Version]
- Ó’Maoiléidigh, D.S.; Wuest, S.E.; Rae, L.; Raganelli, A.; Ryan, P.T.; Kwaśniewska, K.; Das, P.; Lohan, A.J.; Loftus, B.; Graciet, E.; et al. Control of reproductive floral organ identity specification in arabidopsis by the C function regulator AGAMOUS. Plant Cell 2013, 25, 2482–2503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larsson, E.; Roberts, C.J.; Claes, A.R.; Franks, R.G.; Sundberg, E. Polar auxin transport is essential for medial versus lateral tissue specification and vascular-mediated valve outgrowth in Arabidopsis gynoecia. Plant Physiol. 2014, 166, 1998–2012. [Google Scholar] [CrossRef] [Green Version]
- Reyes-Olalde, J.I.; Zuñiga-Mayo, V.M.; Chávez Montes, R.A.; Marsch-Martínez, N.; de Folter, S. Inside the gynoecium: At the carpel margin. Trends Plant Sci. 2013, 18, 644–655. [Google Scholar] [CrossRef]
- Hawkins, C.; Liu, Z. A model for an early role of auxin in Arabidopsis gynoecium morphogenesis. Front. Plant Sci. 2014, 5, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Sehra, B.; Franks, R.G. Auxin and cytokinin act during gynoecial patterning and the development of ovules from the meristematic medial domain. Wiley Interdiscip. Rev. Dev. Biol. 2015, 4, 555–571. [Google Scholar] [CrossRef]
- Marsch-Martínez, N.; de Folter, S. Hormonal control of the development of the gynoecium. Curr. Opin. Plant Biol. 2016, 29, 104–114. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; de Folter, S. Control of stem cell activity in the carpel margin meristem (CMM) in Arabidopsis. Plant Reprod. 2019, 32, 123–136. [Google Scholar] [CrossRef]
- Simonini, S.; Østergaard, L. Female reproductive organ formation: A multitasking endeavor. Curr. Top. Dev. Biol. 2019, 131, 337–371. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Yamaguchi, N.; Gan, E.S.; Ito, T. When to stop: An update on molecular mechanisms of floral meristem termination. J. Exp. Bot. 2019, 70, 1711–1718. [Google Scholar] [CrossRef] [PubMed]
- Skoog, F.; Miller, C.O. Chemical regulation of growth and organ formation in plant tissue cultured in vitro. Symp. Soc. Exp. Biol. 1957, 11, 118–131. [Google Scholar] [PubMed]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucero, L.E.; Uberti-Manassero, N.G.; Arce, A.L.; Colombatti, F.; Alemano, S.G.; Gonzalez, D.H. TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 2015, 84, 267–282. [Google Scholar] [CrossRef]
- Schuster, C.; Gaillochet, C.; Lohmann, J.U. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 2015, 142, 3343–3350. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.J.; Larsson, E.; Spíchal, L.; Sundberg, E. Cytokinin-auxin crosstalk in the gynoecial primordium ensures correct domain patterning. Plant Physiol. 2017, 175, 1144–1157. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; Zúñiga-Mayo, V.M.; Serwatowska, J.; Chavez Montes, R.A.; Lozano-Sotomayor, P.; Herrera-Ubaldo, H.; Gonzalez-Aguilera, K.L.; Ballester, P.; Ripoll, J.J.; Ezquer, I.; et al. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 2017, 13, e1006726. [Google Scholar] [CrossRef]
- Simonini, S.; Bencivenga, S.; Trick, M.; Østergaard, L. Auxin-induced modulation of ETTIN activity orchestrates gene expression in arabidopsis. Plant Cell 2017, 29, 1864–1882. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, N.; Huang, J.; Xu, Y.; Tanoi, K.; Ito, T. Fine-tuning of auxin homeostasis governs the transition from floral stem cell maintenance to gynoecium formation. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef] [Green Version]
- Ó‘Maoiléidigh, D.S.; Stewart, D.; Zheng, B.; Coupland, G.; Wellmer, F. Floral homeotic proteins modulate the genetic program for leaf development to suppress trichome formation in flowers. Development 2018, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rong, X.F.; Sang, Y.L.; Wang, L.; Meng, W.J.; Zou, C.H.; Dong, Y.X.; Bie, X.M.; Cheng, Z.J.; Zhang, X.S. Type-B ARRs control carpel regeneration through mediating AGAMOUS expression in Arabidopsis. Plant Cell Physiol. 2018, 59, 761–769. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Wang, R.; Zi, H.; Li, Y.; Cao, X.; Li, D.; Guo, L.; Tong, J.; Pan, Y.; Jiao, Y.; et al. AUXIN RESPONSE FACTOR3 regulates floral meristem determinacy by repressing cytokinin biosynthesis and signaling. Plant Cell 2018, 30, 324–346. [Google Scholar] [CrossRef] [Green Version]
- Cerbantez-Bueno, V.E.; Zúñiga-Mayo, V.M.; Reyes-Olalde, J.I.; Lozano-Sotomayor, P.; Herrera-Ubaldo, H.; Marsch-Martinez, N.; de Folter, S. Redundant and non-redundant functions of the AHK cytokinin receptors during gynoecium development. Front. Plant Sci. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Venglat, S.P.; Sawhney, V.K. Benzylaminopurine induces phenocopies of floral meristem and organ identity mutants in wild-type Arabidopsis plants. Planta 1996, 198, 480–487. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Nibau, C.; Di Stilio, V.S.; Wu, H.M.; Cheung, A.Y. Arabidopsis and Tobacco SUPERMAN regulate hormone signalling and mediate cell proliferation and differentiation. J. Exp. Bot. 2011, 62, 949–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsch-Martínez, N.; Ramos-Cruz, D.; Irepan Reyes-Olalde, J.; Lozano-Sotomayor, P.; Zúñiga-Mayo, V.M.; de Folter, S. The role of cytokinin during Arabidopsis gynoecia and fruit morphogenesis and patterning. Plant J. 2012, 72, 222–234. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Garcia-Molina, A.; Höhmann, S.; Berndtgen, R.; Huijser, P. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential patterning. Plant J. 2013, 75, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Fernández, I.; Sanchís, S.; Marini, N.; Balanzá, V.; Ballester, P.; Navarrete-Gómez, M.; Oliveira, A.C.; Colombo, L.; Ferrándiz, C. The effect of NGATHA altered activity on auxin signaling pathways within the Arabidopsis gynoecium. Front. Plant Sci. 2014, 5, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Moubayidin, L.; Østergaard, L. Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr. Biol. 2014, 24, 2743–2748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zúñiga-Mayo, V.M.; Reyes-Olalde, J.I.; Marsch-Martinez, N.; de Folter, S. Cytokinin treatments affect the apical-basal patterning of the Arabidopsis gynoecium and resemble the effects of polar auxin transport inhibition. Front. Plant Sci. 2014, 5, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Urbanus, S.L.; de Folter, S.; Shchennikova, A.V.; Kaufmann, K.; Immink, R.G.; Angenent, G.C. In planta localisation patterns of MADS domain proteins during floral development in Arabidopsis thaliana. BMC Plant Biol. 2009, 9, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.H.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schallera, G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 2008, 20, 2102–2116. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Gao, Y.; Zhao, Y. Epigenetic suppression of T-DNA insertion mutants in Arabidopsis. Mol. Plant 2013, 6, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Mason, M.G.; Mathews, D.E.; Argyros, D.A.; Maxwell, B.B.; Kieber, J.J.; Alonso, J.M.; Ecker, J.R.; Schaller, G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 2005, 17, 3007–3018. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, A.; Yamashino, T.; Amano, Y.I.; Tajima, Y.; Imamura, A.; Sakakibara, H.; Mizuno, T. Type-B ARR transcription factors, ARR10 and ARR12, are implicated in cytokinin-mediated regulation of protoxylem differentiation in roots of Arabidopsis thaliana. Plant Cell Physiol. 2007, 48, 84–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishida, K.; Yamashino, T.; Yokoyama, A.; Mizuno, T. Three type-B response regulators, ARR1, ARR10 and ARR12, play essential but redundant roles in cytokinin signal transduction throughout the life cycle of Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 47–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, M.; Chen, H.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 2018, 9, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, N.; Huang, J.; Tatsumi, Y.; Abe, M.; Sugano, S.S.; Kojima, M.; Takebayashi, Y.; Kiba, T.; Yokoyama, R.; Nishitani, K.; et al. Chromatin-mediated feed-forward auxin biosynthesis in floral meristem determinacy. Nat. Commun. 2018, 9, 1–7. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gómez-Felipe, A.; Kierzkowski, D.; de Folter, S. The Relationship between AGAMOUS and Cytokinin Signaling in the Establishment of Carpeloid Features. Plants 2021, 10, 827. https://doi.org/10.3390/plants10050827
Gómez-Felipe A, Kierzkowski D, de Folter S. The Relationship between AGAMOUS and Cytokinin Signaling in the Establishment of Carpeloid Features. Plants. 2021; 10(5):827. https://doi.org/10.3390/plants10050827
Chicago/Turabian StyleGómez-Felipe, Andrea, Daniel Kierzkowski, and Stefan de Folter. 2021. "The Relationship between AGAMOUS and Cytokinin Signaling in the Establishment of Carpeloid Features" Plants 10, no. 5: 827. https://doi.org/10.3390/plants10050827
APA StyleGómez-Felipe, A., Kierzkowski, D., & de Folter, S. (2021). The Relationship between AGAMOUS and Cytokinin Signaling in the Establishment of Carpeloid Features. Plants, 10(5), 827. https://doi.org/10.3390/plants10050827