A Transcriptional Analysis of the Genes Involved in the Ascorbic Acid Pathways Based on a Comparison of the Juice and Leaves of Navel and Anthocyanin-Rich Sweet Orange Varieties
Abstract
:1. Introduction
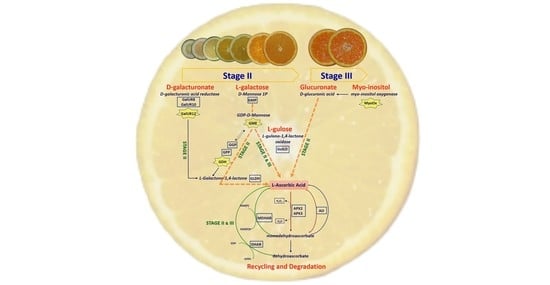
2. Results and Discussion
2.1. AsA in Juice: NH the Genotype with the Highest Content
2.2. Ascorbic Acid Content in the Leaves Is Higher Than That in the Juice
2.3. Expression Analysis of Genes Involved in AsA Biosynthetic Pathways in Juice and Leaves
2.3.1. l-Galactose and l-Gulose Pathways in Juice
GME Plays a Pivotal Role in AsA Biosynthesis
GulLO Contributes to the AsA Content, Mostly during Ripening
GMP, GGP, and GPP
GDH and GLDH: Two Genes That Allow Us to Discriminate NH from Other Varieties
2.3.2. l-Galactose Pathway in the Leaves
2.3.3. GulLO Expression Is Missing in Leaves of Citrus
2.3.4. Myo-Inositol through Glucuronate Pathway in Juice and Leaves
Myo-Inositol in the Juice
Myo-Inositol in the Leaves
2.3.5. d-Galacturonate Pathway
2.3.6. Recycling Pathway
2.3.7. Degradation Pathway
3. Materials and Methods
3.1. Plant Material
3.2. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR Expression Analysis
3.3. Gene Selection and Primer Design
3.4. Chemicals and Physicochemical Analyses
3.4.1. Determination of Ascorbic Acid in Juice
3.4.2. Determination of Ascorbic Acid in Leaves
3.5. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fenech, M.; Amaya, I.; Valpuesta, V.; Botella, M.A. Vitamin C content in fruits: Biosynthesis and regulation. Front. Plant Sci. 2019, 9, 2006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.Y.; Zhang, Q.Y.; Wang, F.; Meng, X.; Meng, Q.W. Ascorbate plays a key role in alleviating low temperature-induced oxidative stress in Arabidopsis. Photosynthetica 2012, 50, 602–612. [Google Scholar] [CrossRef]
- De Tullio, M. Is ascorbic acid a key signaling molecule integrating the activities of 2-oxoglutarate-dependent dioxygenases? Shifting the paradigm. Environ. Exp. Bot. 2020, 178, 104173. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003, 119, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Veljovic-Jovanovic, S.; Foyer, C.H. Peroxide processing in photosynthesis: Antioxidant coupling and redox signalling. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 1465–1475. [Google Scholar] [CrossRef] [Green Version]
- Pignocchi, C.; Foyer, C.H. Apoplastic ascorbate metabolism and its role in the regulation of cell signalling. Curr. Opin. Plant Biol. 2003, 6, 379–389. [Google Scholar] [CrossRef]
- Gallie, D.R. L-ascorbic Acid: A multifunctional molecule supporting plant growth and development. Scientifica 2013, 2013, 795964. [Google Scholar] [CrossRef] [Green Version]
- Xiao, M.; Li, Z.; Zhu, L.; Wang, J.; Zhang, B.; Zheng, F.; Zhao, B.; Zhang, H.; Wang, Y.; Zhang, Z. The Multiple Roles of Ascorbate in the Abiotic Stress Response of Plants: Antioxidant, Cofactor, and Regulator. Front. Plant Sci. 2021, 12, 598173. [Google Scholar] [CrossRef] [PubMed]
- Carr, A.C.; Maggini, S. Vitamin C and immune function. Nutrients 2017, 9, 1211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xylia, P.; Chrysargyris, A.; Tzortzakis, N. The combined and single effect of marjoram essential oil, ascorbic acid, and chitosan on fresh-cut lettuce preservation. Foods 2021, 10, 575. [Google Scholar] [CrossRef]
- Arias, E.; González, J.; Oria, R.; Lopez-Buesa, P. Ascorbic Acid and 4-Hexylresorcinol Effects on Pear PPO and PPO Catalyzed Browning. Reaction. J. Food Sci. 2007, 72, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Cheng, R.Z.; Kogan, M.; Davis, D. Ascorbate as Prophylaxis and Therapy for COVID-19—Update from Shanghai and U.S. Medical Institutions. Glob. Adv. Health Med. 2020, 9, 1–5. [Google Scholar] [CrossRef]
- Cerullo, G.; Negro, M.; Parimbelli, M.; Pecoraro, M.; Perna, S.; Liguori, G.; Rondanelli, M.; Cena, H.; D’Antona, G. The Long History of Vitamin C: From Prevention of the Navel Cold to Potential Aid in the Treatment of COVID-19. Front. Immunol. 2020, 11, 574029. [Google Scholar] [CrossRef] [PubMed]
- Eurostat. Available online: https://ec.europa.eu/eurostat (accessed on 27 April 2021).
- Klimek-Szczykutowicz, M.; Szopa, A.; Ekiert, H. Citrus limon (lemon) phenomenon—A review of the chemistry, pharmacological properties, applications in the modern pharmaceutical, food, and cosmetics industries, and biotechnological studies. Plants 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martí, N.; Mena, P.; Cánovas, J.A.; Micol, V.; Saura, D. Vitamin C and the role of citrus juices as functional food. Nat. Prod. Commun. 2009, 4, 677–700. [Google Scholar] [CrossRef] [Green Version]
- Czech, A.; Malik, A.; Sosnowska, B.; Domaradzki, P. Bioactive substances, heavy metals, and antioxidant activity in whole fruit, peel, and pulp of citrus fruits. Int. J. Food Sci. 2021, 2021, 6662259. [Google Scholar] [CrossRef]
- Gallie, D.R. The role of l-ascorbic acid recycling in responding to environmental stress and in promoting plant growth. J. Exp. Bot. 2013, 64, 433–443. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.Y.; Xie, J.X.; Wang, F.F.; Zhong, J.; Liu, Y.Z.; Li, G.H.; Penga, S.A. Comparison of ascorbate metabolism in fruits of two citrus species with obvious difference in ascorbate content in pulp. J. Plant Physiol. 2011, 168, 2196–2205. [Google Scholar] [CrossRef]
- Linster, C.L.; Clarke, S.G. L-Ascorbate biosynthesis in higher plants: The role of VTC2. Trends Plant Sci. 2008, 13, 567–573. [Google Scholar] [CrossRef] [Green Version]
- Wolucka, B.A.; Van Montagu, M. GDP-mannose 3′,5′-epimerase forms GDP-L-gulose, a putative intermediate for the de novo biosynthesis of vitamin C in plants. J. Biol. Chem. 2003, 278, 47483–47490. [Google Scholar] [CrossRef] [Green Version]
- Running, J.A.; Burlingame, R.P.; Berry, A. The pathway of L-ascorbic acid biosynthesis in the colourless microalga Prototheca moriformis. J. Exp. Bot. 2003, 54, 1841–1849. [Google Scholar] [CrossRef] [Green Version]
- Gilbert, L.; Alhagdow, M.; Nunes-Nesi, A.; Quemener, B.; Guillon, F.; Bouchet, B.; Faurobert, M.; Gouble, B.; Page, D.; Garcia, V.; et al. GDP-D-mannose 3,5-epimerase (GME) plays a key role at the intersection of ascorbate and non-cellulosic cell-wall biosynthesis in tomato. Plant J. 2009, 60, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Agius, F.; González-Lamothe, R.; Caballero, J.L.; MuñozBlanco, J.; Botella, M.A.; Valpuesta, V. Engineering increased vitamin C levels in plants by overexpression of a D-galacturonic acid reductase. Nat. Biotechnol. 2003, 21, 177–181. [Google Scholar] [CrossRef]
- Endres, S.; Tenhaken, R. Myoinositol oxygenase controls the level of myoinositol in Arabidopsis but does not increase ascorbic acid. Plant Physiol. 2009, 149, 1042–1049. [Google Scholar] [CrossRef] [Green Version]
- Endres, S.; Tenhaken, R. Down-regulation of the myo-inositol oxygenase gene family has no effect on cell wall composition in Arabidopsis. Planta 2011, 234, 157–169. [Google Scholar] [CrossRef] [Green Version]
- Duan, J.; Zhang, M.; Zhang, H.; Xiong, H.; Liu, P.; Ali, J.; Li, J.; Li, Z. OsMIOX, a myo-inositol oxygenase gene, improves drought tolerance through scavenging of reactive oxygen species in rice (Oryza sativa L.). Plant Sci. 2012, 196, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Kavkova, E.I.; Blochl, C.; Tenhaken, R. The Myo-inositol pathway does not contribute to ascorbic acid synthesis. Plant Biol. 2018, 21, 95–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorence, A.; Chevone, B.I.; Mendes, P.; Nessler, C.L. Myo-Inositol Oxygenase Offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol. 2004, 134, 1200–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cruz-Rus, E.; Botella, M.A.; Valpuesta, V.; Gomez-Jimenez, M.C. Analysis of genes involved in L-ascorbic acid biosynthesis during growth and ripening of grape berries. J. Plant Physiol. 2010, 167, 739–748. [Google Scholar] [CrossRef]
- Alós, E.; Rodrigo, M.J.; Zacarías, L. Differential transcriptional regulation of l-ascorbic acid content in peel and pulp of citrus fruits during development and maturation. Planta 2014, 239, 1113–1128. [Google Scholar] [CrossRef]
- Mellidou, I.; Keulemans, J.; Kanellis, A.K.; Davey, M.W. Regulation of fruit ascorbic acid concentrations during ripening in high and low vitamin C tomato cultivars. BMC Plant Biol. 2012, 12, 239. [Google Scholar] [CrossRef] [Green Version]
- Chen, Z.; Young, T.E.; Ling, J.; Chang, S.C.; Gallie, D.R. Increasing vitamin C content of plants through enhanced ascorbate recycling. Proc. Natl. Acad. Sci. USA 2003, 100, 3525–3530. [Google Scholar] [CrossRef] [Green Version]
- Hancock, R.D.; Viola, R. Biosynthesis and catabolism of L-ascorbic acid in plants. Crit. Rev. Plant Sci. 2005, 24, 167–188. [Google Scholar] [CrossRef]
- Wang, Z.; Xiao, Y.W.; Tang, K.; Zhang, L. Increased vitamin C content accompanied by an enhanced recycling pathway confers oxidative stress tolerance in Arabidopsis. J. Integr. Plant Biol. 2010, 52, 400–409. [Google Scholar] [CrossRef]
- Qin, A.; Shi, Q.; Yu, X. Ascorbic acid contents in transgenic potato plants overexpressing two dehydroascorbate reductase genes. Mol. Biol. Rep. 2011, 38, 1557–1566. [Google Scholar] [CrossRef] [PubMed]
- Haroldsen, V.M.; Chi-Ham, C.L.; Kulkarni, S.; Lorence, A.; Bennett, A.B. Constitutively expressed DHAR and MDHAR influence fruit, but not foliar ascorbate levels in tomato. Plant Physiol. Biochem. 2011, 49, 1244–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishikawa, T.; Dowdle, J.; Smirnoff, N. Progress in manipulating ascorbic acid biosynthesis and accumulation in plants. Physiol. Plant. 2006, 126, 343–355. [Google Scholar] [CrossRef]
- Badejo, A.A.; Wada, K.; Gao, Y.; Maruta, T.; Sawa, Y.; Shigeoka, S.; Ishikawa, T. Translocation and the alternative D-galacturonate pathway contribute to increasing the ascorbate level in ripening tomato fruits together with the D-mannose/L-galactose pathway. J. Exp. Bot. 2012, 63, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapisarda, P.; Intelisano, S. Sample preparation for vitamin C analysis of pigmented orange juice. Ital. J. Food Sci. 1996, 8, 251–256. [Google Scholar]
- Rapisarda, P.; Lo Bianco, M.; Pannuzzo, P.; Timpanaro, N. Effect of cold storage on vitamin C, phenolics and antioxidant activity of five different orange genotypes [Citrus sinensis (L.) Osbeck]. Postharvest Biol. Technol. 2008, 49, 348–354. [Google Scholar] [CrossRef]
- Bermejo, A.; Cano, A. Analysis of Nutritional Constituents in Twenty Citrus Cultivars from the Mediterranean Area at Different Stages of Ripening. Food Nutr. Sci. 2012, 3, 639–650. [Google Scholar] [CrossRef]
- Cebadera-Miranda, L.; Domínguez, L.; Dias, M.I.; Barros, L.; Ferreira, I.C.F.R.; Igual, M.; Martínez-Navarrete, N.; Fernández-Ruiz, V.; Morales, P.; Cámara, M. Sanguinello and Tarocco (Citrus sinensis [L.] Osbeck): Bioactive compounds and colour appearance of blood oranges. Food Chem. 2019, 270, 395–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Chen, L.L.; Ruan, X.; Chen, D.; Zhu, A.; Chen, C.; Bertrand, D.; Jiao, W.B.; Hao, B.H.; Lyon, M.P.; et al. The Draft Genome of Sweet Orange (Citrus Sinensis). Nat. Genet. 2012, 45, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Lado, J.; Alós, E.; Rodrigo, M.J.; Zacarías, L. Light avoidance reduces ascorbic acid accumulation in the peel of Citrus fruit. Plant Sci. 2015, 231, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Foyer, C.H.; Noctor, G. Ascorbate and Glutathione: The heart of the redox hub. Plant Physiol. 2011, 155, 2–18. [Google Scholar] [CrossRef] [Green Version]
- Bain, J.M. Morphological, anatomical, and physiological changes in the developing fruit of the Valencia orange, Citrus sinensis (L) Osbeck. Aust. J. Bot. 1958, 6, 1–23. [Google Scholar] [CrossRef]
- Gest, N.; Gautier, H.; Stevens, R. Ascorbate as seen through plant evolution: The rise of a successful molecule? J. Exp. Bot. 2013, 64, 33–53. [Google Scholar] [CrossRef] [Green Version]
- Beauvoit, B.; Belouah, I.; Bertin, N.; Cakpo, C.B.; Colombié, S.; Dai, Z.; Gautier, H.; Génard, M.; Moing, A.; Roch, L.; et al. Putting primary metabolism into perspective to obtain better fruits. Ann. Bot. 2018, 122, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Yang, S.X.; Deng, L. Determination of internal qualities of Newhall navel oranges based on NIR spectroscopy using machine learning. J. Food Eng. 2015, 161, 16–23. [Google Scholar] [CrossRef]
- Nagy, S. Vitamin C contents of citrus fruit and their products: A review. J. Agric. Food Chem. 1980, 28, 8–18. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Li, M.; Chen, X.; Wang, P.; Ma, F. Ascorbic Acid Accumulation and Expression of Genes Involved in Its Biosynthesis and Recycling in Developing Apple Fruit. J. Am. Soc. Hortic. Sci. 2011, 136, 231–238. [Google Scholar] [CrossRef] [Green Version]
- Stevens, R.; Buret, M.; Duffe’, P.; Garchery, C.; Baldet, P.; Rothan, C.; Causse, M. Candidate genes and quantitative trait loci affecting fruit ascorbic acid content in three tomato populations. Plant Physiol. 2007, 143, 1943–1953. [Google Scholar] [CrossRef] [Green Version]
- Valpuesta, V.; Botella, M.A. Biosynthesis of L-ascorbic acid in plants: New pathways for an old antioxidant. Trends Plant Sci. 2004, 9, 573–577. [Google Scholar] [CrossRef] [PubMed]
- Wolucka, B.A.; Van Montagu, M. The VTC2 cycle and the de novo biosynthesis pathways for vitamin C in plants: An opinion. Phytochemistry 2007, 68, 2602–2613. [Google Scholar] [CrossRef] [PubMed]
- Ioannidi, E.; Kalamaki, M.S.; Engineer, C.; Pateraki, I.; Alexandrou, D.; Mellidou, I.; Giovannonni, J.; Kanellis, A.K. Expression profiling of ascorbic acid-related genes during tomato fruit development and ripening and in response to stress conditions. J. Exp. Bot. 2009, 60, 663–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bulley, S.M.; Rassam, M.; Hoser, D.; Otto, W.; Schunemann, N.; Wright, M.; MacRae, E.; Gleave, A.; Laing, W. Gene expression studies in kiwifruit and gene over-expression in Arabidopsis indicates that GDP-L-galactose guanyltransferase is a major control point of vitamin C biosynthesis. J. Exp. Bot. 2009, 60, 765–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yungyuen, W.; Ma, G.; Zhang, L.; Yamawaki, K.; Yahata, M.; Ohta, S.; Yoshioka, T.; Kato, M. Regulation of ascorbic acid metabolism in response to different temperatures in citrus juice sacs in vitro. Sci. Hortic. 2017, 217, 1–7. [Google Scholar] [CrossRef]
- Dowdle, J.; Ishikawa, T.; Gatzek, S.; Rolinski, S.; Smirnoff, N. Two genes in Arabidopsis thaliana encoding GDP-L-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J. 2007, 52, 673–689. [Google Scholar] [CrossRef]
- Laing, W.A.; Wright, M.A.; Cooney, J.; Bulley, S.M. The missing step of the L-galactose pathway of ascorbate biosynthesis in plants, an L-galactose guanyltransferase, increases leaf ascorbate content. Proc. Natl. Acad. Sci. USA 2007, 104, 9534–9539. [Google Scholar] [CrossRef] [Green Version]
- Ye, W.; Ren, W.; Kong, L.; Zhang, W.; Wang, T. Transcriptomic profiling analysis of Arabidopsis thaliana treated with exogenous Myo-Inositol. PLoS ONE 2016, 11, e0161949. [Google Scholar] [CrossRef]
- Kanter, U.; Usadel, B.; Guerineau, F.; Li, Y.; Pauly, M.; Tenhaken, R. The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 2005, 221, 243–254. [Google Scholar] [CrossRef] [Green Version]
- Smirnoff, N.; Conklin, P.L.; Loewus, F.A. Biosynthesis of ascorbic acid in plants: A renaissance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 437–467. [Google Scholar] [CrossRef] [PubMed]
- Chiaiese, P.; Corrado, G.; Minutolo, M.; Barone, A.; Errico, A. Transcriptional Regulation of Ascorbic Acid During Fruit Ripening in Pepper (Capsicum annuum) Varieties with Low and High Antioxidants Content. Plants 2019, 8, 206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alós, E.; Rodrigo, M.J.; Zacarías, L. Transcriptomic analysis of genes involved in the biosynthesis, recycling and degradation of L-ascorbic acid in pepper fruits (Capsicum annuum L.) degradation of L-ascorbic acid in pepper fruits (Capsicum annuum L.). Plant Sci. 2013, 207, 2–11. [Google Scholar] [CrossRef]
- Laing, W.A.; Bulley, S.; Wright, M.; Cooney, J.; Jensen, D.; Barraclough, D.; MacRae, E. A highly specific L-galactose-1-phosphate phosphatase on the path to ascorbate biosynthesis. Proc. Natl. Acad. Sci. USA 2004, 101, 16976–16981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torabinejad, J.; Donahue, J.L.; Gunesekera, B.N.; Allen-Daniels, M.J.; Gillaspy, G.E. VTC4 is a bifunctional enzyme that affects myo-inositol and ascorbate biosynthesis in plants. Plant Physiol. 2009, 150, 951–961. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Rus, E.; Amaya, I.; Sánchez-Sevilla, J.F.; Botella, M.A.; Valpuesta, V. Regulation of L-ascorbic acid content in strawberry fruits. J. Exp. Bot. 2011, 62, 4191–4201. [Google Scholar] [CrossRef]
- Aboobucker, S.I.; Suza, W.P.; Lorence, A. Characterization of Two Arabidopsis L-Gulono-1,4-lactone Oxidases, AtGulLO3 and AtGulLO5, Involved in Ascorbate Biosynthesis. React. Oxyg. Species 2017, 4, 389–417. [Google Scholar] [CrossRef]
- Munir, S.; Muhammad, A.M.; Ahiakpa, J.K.; Liu, G.; Chen, W.; Zhou, G.; Zheng, W.; Ye, Z.; Zhang, Y. Genome-wide analysis of Myo-inositol oxygenase gene family in tomato reveals their involvement in ascorbic acid accumulation. BMC Genom. 2020, 21, 284. [Google Scholar] [CrossRef] [Green Version]
- Mellidou, I.; Kanellis, A.K. Genetic control of ascorbic acid biosynthesis and recycling in horticultural crops. Front. Chem. 2017, 5, 50. [Google Scholar] [CrossRef] [Green Version]
- Noctor, G.; Foyer, C.H. Ascorbate and glutathione: Keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 249–279. [Google Scholar] [CrossRef] [PubMed]
- Eltayeb, A.E.; Naoyoshi, K.; Badawi, G.H.; Kaminaka, H.; Sanekata, T.; Shibahara, T.; Inanaga, S.; Tanaka, K. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta 2007, 225, 1255–1264. [Google Scholar] [CrossRef]
- Stevens, R.; Page, D.; Gouble, B.; Garchery, C.; Zamir, D.; Causse, M. Tomato fruit ascorbic acid content is linked with monodehydroascorbate reductase activity and tolerance to chilling stress. Plant Cell Environ. 2008, 31, 1086–1096. [Google Scholar] [CrossRef]
- El Airaj, H.; Gest, N.; Truffault, V.; Garchery, C.; Riqueau, G.; Gouble, B.; Page, D.; Stevens, R. Decreased monodehydroascorbate reductase activity reduces tolerance to cold storage in tomato and affects fruit antioxidant levels. Postharvest Biol. Technol. 2013, 86, 502–510. [Google Scholar] [CrossRef]
- Maruta, T.; Sawa, Y.; Shigeoka, S.; Ishikawa, T. Diversity and evolution of ascorbate peroxidase functions in chloroplasts: More than just a classical antioxidant enzyme? Plant Cell Physiol. 2016, 57, 1377–1386. [Google Scholar] [CrossRef] [Green Version]
- De Tullio, M.C.; Liso, R.; Arrigoni, O. Ascorbic Acid Oxidase: An Enzyme in Search of a Role. Biol. Plant. 2004, 48, 161–166. [Google Scholar] [CrossRef]
- Sanmartin, M.; Pateraki, I.; Chatzopoulou, F.; Kanellis, A.K. Differential expression of the ascorbate oxidase multigene family during fruit development and in response to stress. Planta 2007, 225, 873–885. [Google Scholar] [CrossRef] [PubMed]
- Mazid, M.; Khan, T.A.; Khan, Z.H.; Quddusi, S.; Mohammad, F. Occurrence, biosynthesis and potentialities of ascorbic acid in plants. Int. J. Plant Anim. Environ. Sci. 2011, 1, 167–184. [Google Scholar]
- De Tullio, M.C.; Ciraci, S.; Liso, R.; Arrigoni, O. Ascorbic acid oxidase is dynamically regulated by light and oxygen. A tool for oxygen management in plants? J. Plant Physiol. 2005, 164, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Arrigoni, O.; Chinni, E.; Ciraci, S.; De Tullio, M.C. In vivo elicitation of ascorbate oxidase activity by dioxygen and its possible role in photosynthesizing leaves. Rend. Fis. Acc. Lincei 2003, 14, 127–134. [Google Scholar] [CrossRef]
- Zhang, L.; Ma, G.; Yamawaki, K.; Ikoma, Y.; Matsumoto, H.; Yoshioka, T.; Ohta, S.; Kato, M. Regulation of ascorbic acid metabolism by blue LED light irradiation in citrus juice sacs. Plant Sci. 2015, 233, 134–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mastropasqua, L.; Borraccino, G.; Bianco, L.; Paciolla, C. Light qualities and dose influence ascorbate pool size in detached oat leaves. Plant Sci. 2011, 183, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Caruso, M.; Ferlito, F.; Licciardello, C.; Allegra, M.; Strano, M.C.; Di Silvestro, S.; Russo, M.P.; Paolo, D.P.; Caruso, P.; Las Casas, G.; et al. Pomological diversity of the Italian blood orange germplasm. Sci. Hortic. 2016, 213, 331–339. [Google Scholar] [CrossRef]
- Barry, G.H.; Caruso, M.; Gmitter, F.G., Jr. Commercial scion varieties. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Eds.; Woodhead Publishing: Duxford, UK, 2020; pp. 83–104. [Google Scholar]
- Bernardi, J.; Licciardello, C.; Russo, M.P.; Chiusano, M.L.; Carletti, G.; Reforgiato Recupero, G.; Marocco, A. Use of a custom array to study differential expressed genes during pigmented orange (Citrus sinensis L. Osbeck) ripening. J. Plant Physiol. 2010, 167, 301–310. [Google Scholar] [CrossRef]
- Mahe, A.; Grisvard, J.; Dron, M. Fungal-and plant-specific gene markers to follow the bean anthracnose infection process and normalize a bean chitinase mRNA induction. Mol. Plant Microbe Interact. 1992, 5, 242–248. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A Better Web Interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef]
- Kõressaar, T.; Lepamets, M.; Kaplinski, L.; Raime, K.; Andreson, R.; Remm, M. Primer3-Masker: Integrating Masking of Template Sequence with Primer Design Software. Bioinformatics 2018, 34, 1937–1938. [Google Scholar] [CrossRef]
- Kimball, D. Citrus Processing: Quality Control and Technology; AVI Book: New York, NY, USA, 1991; p. 473. [Google Scholar]
- Zhong, C.F.; Mazzoni, L.; Balducci, F.; Di Vittori, L.; Capocasa, F.; Giampieri, F.; Mezzetti, B. Evaluation of vitamin C content in fruit and leaves of different strawberry genotypes. Acta Hortic. 2017, 1156, 371–377. [Google Scholar] [CrossRef]
- Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. Available online: https://rpkgs.datanovia.com/factoextra/ (accessed on 27 April 2021).
- Fellows, I. A data analysis GUI for r. J. Stat. Softw. 2012, 49, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis. 2016. Available online: https://ggplot2.tidyverse.org (accessed on 26 April 2021).













| Juice | Leaves | |||||||
|---|---|---|---|---|---|---|---|---|
| NH | LL | TDV | TM | NH | LL | TDV | TM | |
| GMP | −0.22 | −0.55 | −0.33 | −0.58 | −0.12 | 0.19 | −0.12 | −0.30 |
| GME | 0.52 | −0.11 | 0.56 | 0.52 | −0.68 | −0.29 | −0.08 | −0.37 |
| GGP | −0.30 | −0.64 | −0.03 | −0.59 | −0.54 | −0.37 | −0.35 | −0.58 |
| GPP | −0.04 | −0.54 | −0.18 | −0.52 | −0.56 | −0.27 | 0.26 | −0.23 |
| GDH | 0.76 | 0.84 | 0.73 | 0.66 | −0.64 | −0.10 | 0.09 | −0.59 |
| GLDH | 0.56 | 0.27 | −0.35 | −0.02 | −0.30 | 0.44 | −0.34 | −0.14 |
| GulLO | −0.61 | −0.54 | −0.65 | −0.81 | n.d. | n.d. | n.d. | n.d. |
| MyoIFS | 0.18 | −0.11 | −0.04 | 0.19 | −0.63 | −0.55 | −0.45 | −0.43 |
| MyoOx | 0.79 | 0.85 | 0.88 | 0.80 | 0.05 | −0.16 | −0.28 | 0.37 |
| MDHAR | −0.46 | −0.67 | 0.38 | −0.84 | −0.59 | 0.10 | −0.25 | −0.42 |
| DHAR | 0.58 | −0.50 | 0.49 | −0.03 | −0.59 | −0.22 | −0.57 | −0.30 |
| GalUR8 | −0.75 | −0.60 | −0.65 | −0.56 | −0.34 | 0.47 | −0.58 | −0.25 |
| GalUR10 | 0.23 | 0.20 | 0.27 | 0.33 | −0.58 | 0.68 | 0.23 | 0.08 |
| GalUR12 | 0.74 | 0.16 | 0.11 | 0.56 | −0.52 | −0.24 | −0.71 | −0.61 |
| APX2 | 0.03 | −0.51 | −0.07 | −0.11 | −0.32 | 0.45 | 0.37 | 0.02 |
| APX3 | −0.31 | −0.49 | −0.17 | 0.03 | −0.75 | 0.50 | 0.16 | −0.27 |
| AO | 0.90 | 0.90 | 0.93 | 0.84 | −0.11 | −0.35 | 0.26 | −0.46 |
| TA | pH | TSS | TSS/TA | AsA | |
|---|---|---|---|---|---|
| August | |||||
| NH | 3.07 | 2.88 | 11.26 | 3.59 | 89.20 |
| LL | 4.35 | 2.74 | 11.03 | 2.53 | 85.00 |
| TDV | 4.48 | 2.76 | 10.83 | 2.42 | 88.14 |
| TM | 5.99 | 2.63 | 10.56 | 1.76 | 80.18 |
| September | |||||
| NH | 2.32 | 3.07 | 12.21 | 5.26 | 79.45 |
| LL | 3.36 | 2.92 | 10.20 | 3.03 | 68.31 |
| TDV | 3.87 | 2.93 | 9.60 | 2.48 | 69.37 |
| TM | 5.19 | 2.72 | 10.00 | 1.94 | 64.87 |
| October | |||||
| NH | 2.46 | 3.16 | 11.22 | 4.56 | 61.70 |
| LL | 2.62 | 3.23 | 8.85 | 3.39 | 54.99 |
| TDV | 3.15 | 3.13 | 9.46 | 3.00 | 57.90 |
| TM | 4.48 | 2.97 | 9.21 | 2.06 | 65.16 |
| November | |||||
| NH | 1.54 | 3.15 | 11.83 | 7.76 | 67.93 |
| LL | 1.77 | 3.06 | 8.57 | 4.84 | 56.89 |
| TDV | 2.02 | 3.01 | 9.45 | 4.69 | 61.74 |
| TM | 3.19 | 2.76 | 9.24 | 2.90 | 61.50 |
| December | |||||
| NH | 1.25 | 3.38 | 12.01 | 9.74 | 61.81 |
| LL | 1.49 | 3.24 | 9.47 | 6.39 | 47.03 |
| TDV | 1.61 | 3.18 | 9.61 | 6.00 | 53.76 |
| TM | 2.70 | 2.86 | 9.34 | 3.47 | 46.57 |
| January | |||||
| NH | 1.13 | 3.38 | 12.17 | 10.87 | 62.12 |
| LL | 1.45 | 3.27 | 9.64 | 6.83 | 47.95 |
| TDV | 1.75 | 3.24 | 9.64 | 5.52 | 50.55 |
| TM | 2.72 | 2.84 | 9.30 | 3.42 | 49.92 |
| February | |||||
| NH | 0.86 | 3.55 | 13.04 | 16.58 | 60.18 |
| LL | 1.04 | 3.41 | 10.61 | 10.26 | 53.10 |
| TDV | 0.98 | 3.51 | 10.21 | 10.43 | 53.38 |
| TM | 2.02 | 2.99 | 10.59 | 6.60 | 52.98 |
| March | |||||
| NH | 0.88 | 3.63 | 12.00 | 14.06 | 60.40 |
| LL | 0.96 | 3.56 | 10.50 | 10.96 | 47.63 |
| TDV | 1.11 | 3.47 | 9.78 | 8.81 | 51.63 |
| TM | 1.62 | 2.96 | 10.22 | 5.09 | 50.71 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Caruso, P.; Russo, M.P.; Caruso, M.; Guardo, M.D.; Russo, G.; Fabroni, S.; Timpanaro, N.; Licciardello, C. A Transcriptional Analysis of the Genes Involved in the Ascorbic Acid Pathways Based on a Comparison of the Juice and Leaves of Navel and Anthocyanin-Rich Sweet Orange Varieties. Plants 2021, 10, 1291. https://doi.org/10.3390/plants10071291
Caruso P, Russo MP, Caruso M, Guardo MD, Russo G, Fabroni S, Timpanaro N, Licciardello C. A Transcriptional Analysis of the Genes Involved in the Ascorbic Acid Pathways Based on a Comparison of the Juice and Leaves of Navel and Anthocyanin-Rich Sweet Orange Varieties. Plants. 2021; 10(7):1291. https://doi.org/10.3390/plants10071291
Chicago/Turabian StyleCaruso, Paola, Maria Patrizia Russo, Marco Caruso, Mario Di Guardo, Giuseppe Russo, Simona Fabroni, Nicolina Timpanaro, and Concetta Licciardello. 2021. "A Transcriptional Analysis of the Genes Involved in the Ascorbic Acid Pathways Based on a Comparison of the Juice and Leaves of Navel and Anthocyanin-Rich Sweet Orange Varieties" Plants 10, no. 7: 1291. https://doi.org/10.3390/plants10071291
APA StyleCaruso, P., Russo, M. P., Caruso, M., Guardo, M. D., Russo, G., Fabroni, S., Timpanaro, N., & Licciardello, C. (2021). A Transcriptional Analysis of the Genes Involved in the Ascorbic Acid Pathways Based on a Comparison of the Juice and Leaves of Navel and Anthocyanin-Rich Sweet Orange Varieties. Plants, 10(7), 1291. https://doi.org/10.3390/plants10071291