Identification and Characterization of Arbutus unedo L. Endophytic Bacteria Isolated from Wild and Cultivated Trees for the Biological Control of Phytophthora cinnamomi
Abstract
:1. Introduction
2. Results
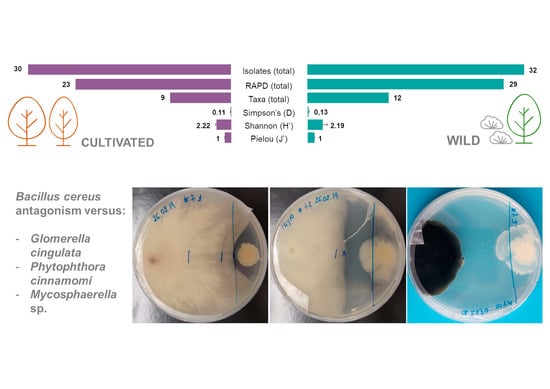
2.1. Isolation of Endophytic Bacteria from Cultivated and Wild Strawberry Trees
2.2. Antagonism Effect of Endophytic Bacteria against Plant Pathogens
2.3. Plant Growth-Promoting Potential of Endophytic Bacteria Isolated from Strawberry Tree
3. Discussion
4. Materials and Methods
4.1. Collection of Plant Material
4.2. Isolation of Endophytic Bacteria
4.3. Identification of Endophytic Bacteria
4.3.1. DNA Extraction and RAPD Fingerprint
4.3.2. Phylogenetic Analysis
4.4. Antagonism Ability of Bacterial Endophytes against A. unedo Fungal Pathogens
4.5. Plant Growth-Promoting Potential of Endophytic Bacteria
4.5.1. Siderophores Production
4.5.2. Phosphate Solubilization
4.5.3. Ammonia Production
4.5.4. Indole-3-Acetic Acid Production
4.6. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Torres, J.A.; Valle, F.; Pinto, C.; García-Fuentes, A.; Salazar, C.; Cano, E. Arbutus unedo L. communities in southern Iberian Peninsula mountains. Plant Ecol. 2002, 160, 207–223. [Google Scholar] [CrossRef]
- Martins, J.; Monteiro, P.; Pinto, G.; Canhoto, J. Hybridization assays in strawberry tree toward the identification of plants displaying increased drought tolerance. Forests 2021, 12, 148. [Google Scholar] [CrossRef]
- Martins, J.F.; Correia, S.I.; Canhoto, J.M. Somatic embryogenesis induction and plant regeneration in strawberry tree (Arbutus unedo L.). In Methods in Molecular Biology; Germana, M., Lambardi, M., Eds.; Humana Press: New York, NY, USA, 2016; Volume 1359, pp. 329–339. [Google Scholar]
- Martins, J.F.; Santos, T.; Correia, S.I.; Canhoto, J.M. Somatic embryogenesis in Arbutus unedo L. and other Ericaceae. In Vegetative Propagation of Forest Trees; Park, Y.S., Bonga, J.M., Moon, H.K., Eds.; National Institute of Forest Science: Seoul, Korea, 2016; pp. 565–590. [Google Scholar]
- Morgado, S.; Morgado, M.; Plácido, A.I.; Roque, F.; Duarte, A.P. Arbutus unedo L.: From traditional medicine to potential uses in modern pharmacotherapy. J. Ethnopharmacol. 2018, 225, 90–102. [Google Scholar] [CrossRef] [PubMed]
- Polizzi, G.; Aiello, D.; Guarnaccia, V.; Vitale, A.; Perrone, G.; Stea, G. First report of damping-off on strawberry tree caused by Colletotrichum acutatum and C. simmondsii in Italy. Plant Dis. 2011, 95, 1588. [Google Scholar] [CrossRef] [PubMed]
- Romero-Martin, M.A.; Trapero-Casas, A. La mancha foliar del madroño (Arbutus unedo) causada por Septoria unedonis var. vellanensis. Bol. Sanid. Veg. Plagas 2003, 29, 375–392. [Google Scholar]
- Jung, T.; Pérez-Sierra, A.; Durán, A.; Jung, M.H.; Balci, Y.; Scanu, B. Canker and decline diseases caused by soil- and airborne Phytophthora species in forests and woodlands. Pers. Mol. Phylogeny Evol. Fungi 2018, 40, 182–220. [Google Scholar] [CrossRef] [Green Version]
- Sena, K.; Crocker, E.; Vincelli, P.; Barton, C. Phytophthora cinnamomi as a driver of forest change: Implications for conservation and management. For. Ecol. Manag. 2018, 409, 799–807. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Whipps, J.M.; Hand, P.; Pink, D.; Bending, G.D. Phyllosphere microbiology with special reference to diversity and plant genotype. J. Appl. Microbiol. 2008, 105, 1744–1755. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Liu, P.; Ye, W. Illumina-based analysis of endophytic bacterial diversity of tree peony (Paeonia Sect. Moutan) roots and leaves. Braz. J. Microbiol. 2017, 48, 695–705. [Google Scholar] [CrossRef]
- Zinniel, D.K.; Lambrecht, P.; Harris, N.B.; Feng, Z.; Kuczmarski, D.; Higley, P.; Ishimaru, C.A.; Arunakumari, A.; Barletta, R.G.; Vidaver, A.K. Isolation and characterization of endophytic colonizing bacteria from agronomic crops and prairie plants. Appl. Environ. Microbiol. 2002, 68, 2198–2208. [Google Scholar] [CrossRef] [Green Version]
- Rabiey, M.; Hailey, L.E.; Roy, S.R.; Grenz, K.; Al-Zadjali, M.A.S.; Barrett, G.A.; Jackson, R.W. Endophytes vs tree pathogens and pests: Can they be used as biological control agents to improve tree health? Eur. J. Plant Pathol. 2019, 155, 711–729. [Google Scholar] [CrossRef] [Green Version]
- Toju, H.; Okayasu, K.; Notaguchi, M. Leaf-associated microbiomes of grafted tomato plants. Sci. Rep. 2019, 9, 1787. [Google Scholar] [CrossRef]
- Wassermann, B.; Müller, H.; Berg, G. An apple a day: Which bacteria do we eat with organic and conventional apples? Front. Microbiol. 2019, 10, 1629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lopes, R.; Tsui, S.; Gonçalves, P.J.R.O.; de Queiroz, M.V. A look into a multifunctional toolbox: Endophytic Bacillus species provide broad and underexploited benefits for plants. World J. Microbiol. Biotechnol. 2018, 34, 94. [Google Scholar] [CrossRef] [PubMed]
- Rocha, F.Y.O.; de Oliveira, C.M.; da Silva, P.R.A.; de Melo, L.H.V.; do Carmo, M.G.F.; Baldani, J.I. Taxonomical and functional characterization of Bacillus strains isolated from tomato plants and their biocontrol activity against races 1, 2 and 3 of Fusarium oxysporum f. sp. Lycopersici. Appl. Soil Ecol. 2017, 120, 8–19. [Google Scholar] [CrossRef]
- Sun, H.; He, Y.; Xiao, Q.; Ye, R.; Tian, Y. Isolation, characterization, and antimicrobial activity of endophytic bacteria from Polygonum cuspidatum. Afr. J. Microbiol. Res. 2013, 7, 1496–1504. [Google Scholar] [CrossRef] [Green Version]
- Varma, P.K.; Kumar, K.V.K.; Sekhar, V.C.; Adilakshmi, D.; Suresh, M.; Kumar, N.R.; Jayachandra, K.; Anitha, R. Evaluation of endophytic bacteria for plant growth promotion and pathogen suppression traits in Saccharum officinarum. J. Agric. Sci. Technol. A 2017, 7, 537–545. [Google Scholar] [CrossRef]
- Khaksar, G.; Treesubsuntorn, C.; Thiravetyan, P. Effect of endophytic Bacillus cereus ERBP inoculation into non-native host: Potentials and challenges for airborne formaldehyde removal. Plant Physiol. Biochem. 2016, 107, 326–336. [Google Scholar] [CrossRef]
- Hu, H.J.; Chen, Y.L.; Wang, Y.F.; Tang, Y.Y.; Chen, S.L.; Yan, S.Z. Endophytic Bacillus cereus effectively controls Meloidogyne incognita on tomato plants through rapid rhizosphere occupation and repellent action. Plant Dis. 2017, 101, 448–455. [Google Scholar] [CrossRef] [Green Version]
- Dahmani, M.A.; Desrut, A.; Moumen, B.; Verdon, J.; Mermouri, L.; Kacem, M.; Coutos-Thévenot, P.; Kaid-Harche, M.; Bergès, T.; Vriet, C. Unearthing the plant growth-promoting traits of Bacillus megaterium RmBm31, an endophytic bacterium isolated from root nodules of Retama monosperma. Front. Plant Sci. 2020, 11, 124. [Google Scholar] [CrossRef]
- Ferreira, A.; Quecine, M.C.; Lacava, P.T.; Oda, S.; Azevedo, J.L.; Araújo, W.L. Diversity of endophytic bacteria from Eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 2008, 287, 8–14. [Google Scholar] [CrossRef] [Green Version]
- McInroy, J.A.; Kloepper, J.W. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 1995, 173, 337–342. [Google Scholar] [CrossRef]
- Pandya, M.; Rajput, M.; Rajkumar, S. Exploring plant growth promoting potential of non rhizobial root nodules endophytes of Vigna radiata. Microbiology 2015, 84, 80–89. [Google Scholar] [CrossRef]
- Long, H.H.; Sonntag, D.G.; Schmidt, D.D.; Baldwin, I.T. The structure of the culturable root bacterial endophyte community of Nicotiana attenuata is organized by soil composition and host plant ethylene production and perception. New Phytol. 2010, 185, 554–567. [Google Scholar] [CrossRef] [PubMed]
- Rong, S.; Xu, H.; Li, L.; Chen, R.; Gao, X.; Xu, Z. Antifungal activity of endophytic Bacillus safensis B21 and its potential application as a biopesticide to control rice blast. Pestic. Biochem. Physiol. 2020, 162, 69–77. [Google Scholar] [CrossRef]
- Sekhar, A.C.; Thomas, P. Isolation and identification of shoot-Tip associated endophytic bacteria from Banana cv. Grand Naine and testing for antagonistic activity against Fusarium oxysporum f. sp. cubense. Am. J. Plant Sci. 2015, 6, 943–954. [Google Scholar] [CrossRef] [Green Version]
- Wu, T.; Xu, J.; Liu, J.; Guo, W.H.; Li, X.B.; Xia, J.B.; Xie, W.J.; Yao, Z.G.; Zhang, Y.M.; Wang, R.Q. Characterization and initial application of endophytic Bacillus safensis strain ZY16 for improving phytoremediation of oil-contaminated saline soils. Front. Microbiol. 2019, 10, 991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuo, L.; Liu, F.; Yan, X.R.; Liu, Y. Bacillus taxi sp. nov., a novel endophytic bacterium isolated from root of Taxus chinensis (Pilger) Rehd. Int. J. Syst. Evol. Microbiol. 2020, 70, 481–486. [Google Scholar] [CrossRef]
- Zineb, F.B.; Chahinez, M.; Abdelkader, B.; Sonia, S.; Odile, D.; Robin, D.; Antoine, G. Nodular bacterial endophyte diversity associated with native Acacia spp. in desert region of Algeria. Afr. J. Microbiol. Res. 2016, 10, 634–645. [Google Scholar] [CrossRef] [Green Version]
- Guo, G.N.; Zhou, X.; Chen, Z.L.; Yang, Z.W.; Li, X.D.; Li, Y.H. Paenibacillus marchantiophytorum sp. nov., isolated from the liverwort Herbertus sendtneri. Int. J. Syst. Evol. Microbiol. 2016, 66, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Niem, J.M.; Billones-Baaijens, R.; Stodart, B.; Savocchia, S. Diversity profiling of grapevine microbial endosphere and antagonistic potential of endophytic Pseudomonas against grapevine trunk diseases. Front. Microbiol. 2020, 11, 477. [Google Scholar] [CrossRef]
- Nawangsih, A.A.; Damayanti, I.; Wiyono, S.; Kartika, J.G. Selection and characterization of endophytic bacteria as biocontrol agents of tomato bacterial wilt disease. J. Biosci. 2011, 18, 66–70. [Google Scholar] [CrossRef] [Green Version]
- Loreti, S.; Gallelli, A.; de Simone, D.; Bosco, A. Detection of Pseudomonas avellanae and the bacterial microflora of hazelnut affected by “Moria” in central Italy. J. Plant Pathol. 2009, 91, 365–373. [Google Scholar] [CrossRef]
- Scortichini, M.; Natalini, E.; Marchesi, U. Evidence for separate origins of the two Pseudomonas avellanae lineages. Plant Pathol. 2006, 55, 451–457. [Google Scholar] [CrossRef]
- Manching, H.C.; Balint-Kurti, P.J.; Stapleton, A.E. Southern leaf blight disease severity is correlated with decreased maize leaf epiphytic bacterial species richness and the phyllosphere bacterial diversity decline is enhanced by nitrogen fertilization. Front. Plant Sci. 2014, 5, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Microbiol. 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of plant growth-promoting bacteria for biocontrol of plant diseases: Principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y. Auxin biosynthesis and its role in plant development. Annu. Rev. Plant Biol. 2010, 61, 49–64. [Google Scholar] [CrossRef] [Green Version]
- Wagi, S.; Ahmed, A. Bacillus spp.: Potent microfactories of bacterial IAA. PeerJ 2019, 7, e7258. [Google Scholar] [CrossRef] [Green Version]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witzell, J.; Martín, J.A. Endophytes and forest health. In Endophytes of Forest Trees; Pirttila, A., Frank, A., Eds.; Springer International Publishing AG: New York, NY, USA, 2018; Volume 86, pp. 261–282. [Google Scholar]
- Krabel, D.; Morgenstern, K.; Herzog, S. Endophytes in changing environments—Do we need new concepts in forest management? iForest 2013, 6, 112. [Google Scholar] [CrossRef]
- Kubiak, K.; Wrzosek, M.; Przemieniecki, S.; Damszel, M.; Sierota, Z. Bacteria inhabiting wood of roots and stumps in forest and arable soils. In Endophytes of Forest Trees; Pirttila, A., Frank, A., Eds.; Springer International Publishing AG: New York, NY, USA, 2018; Volume 86, pp. 319–342. [Google Scholar]
- Eevers, N.; Gielen, M.; Sánchez-López, A.; Jaspers, S.; White, J.C.; Vangronsveld, J.; Weyens, N. Optimization of isolation and cultivation of bacterial endophytes through addition of plant extract to nutrient media. Microb. Biotechnol. 2015, 8, 707–715. [Google Scholar] [CrossRef]
- Tiago, I.; Chung, A.P.; Veríssimo, A. Bacterial diversity in a nonsaline alkaline environment: Heterotrophic aerobic populations. Appl. Environ. Microbiol. 2004, 70, 7378–7387. [Google Scholar] [CrossRef] [Green Version]
- Mergeay, M.; Nies, D.; Schlegel, H.G.; Gerits, J.; Charles, P.; Van Gijsegem, F. Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals. J. Bacteriol. 1985, 162, 328–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedmann-Al-Ahmad, M.; Tichy, H.V.; Schon, G. Characterization of Acinetobacter type strains and isolates obtained from wastewater treatment plants by PCR fingerprinting. Appl. Environ. Microbiol. 1994, 60, 4066–4071. [Google Scholar] [CrossRef] [Green Version]
- Costa, J.; Tiago, I.; Da Costa, M.S.; Veríssimo, A. Presence and persistence of Legionella spp. in groundwater. Appl. Environ. Microbiol. 2005, 71, 663–671. [Google Scholar] [CrossRef] [Green Version]
- Rainey, F.A.; Ward-Rainey, N.; Kroppenstedt, R.M.; Stackebrandt, E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: Proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 1996, 46, 1088–1092. [Google Scholar] [CrossRef]
- Turner, S.; Pryer, K.M.; Miao, V.P.W.; Palmer, J.D. Investigating deep phylogenetic relationships among cyanobacteria and plastids by small subunit rRNA sequence analysis. J. Eukaryot. Microbiol. 1999, 46, 327–338. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Thompson, J.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.; Kishino, H.; Yano, T. Dating of the human-ape splitting by a molecular clock of mitochondrial DNA. J. Mol. Evol. 1985, 22, 160–174. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef] [Green Version]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A.G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dennis, C.; Webster, J. Antagonistic properties of species-groups of Trichoderma: III. Hyphal interaction. Trans. Br. Mycol. Soc. 1971, 57, 363–369. [Google Scholar] [CrossRef]
- Lahlali, R.; Bajii, M.; Jijakli, M.H. Isolation and evaluation of bacteria and fungi as biological control agents against Rhizoctonia solani. Commun. Agric. Appl. Biol. Sci. 2007, 72, 973–982. [Google Scholar]
- Almoneafy, A.A.; Xie, G.L.; Tian, W.X.; Xu, L.H.; Zhang, G.Q.; Ibrahim, M. Characterization and evaluation of Bacillus isolates for their potential plant growth and biocontrol activities against tomato bacterial wilt. Afr. J. Biotechnol. 2012, 11, 7193–7201. [Google Scholar] [CrossRef]
- Schwyn, B.; Neilands, J.B. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Singh, D.; Kumar Yadav, D. Potential of Bacillus amyloliquefaciens for biocontrol of bacterial wilt of tomato incited by Ralstonia solanacearum. J. Plant. Pathol. Microbiol. 2016, 7, 1–6. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 1 July 2020).
- Chen, H. VennDiagram: Generate High-Resolution Venn and Euler Plots. R Package Version 1.6.20. 2018. Available online: https://CRAN.R-project.org/package=VennDiagram (accessed on 1 July 2020).






| Bacteria Endophyte | Plant Species |
|---|---|
| Bacillus cereus | Teucrium polium and Sophora alopecuroides [17], Solanum lycopersicum [18], Polygonum cuspidatum [19], Saccharum officinarum [20], Clitoria ternatea [21], Fragraria ananassa and Dyospiros kaki [22] |
| B. megaterium | Retama monosperma [23], Eucalyptus spp. [24], Gossypium hirsutum and Zea mays [25], S. lycopersicum [18], S. officinarum [20] and V. radiata [26] |
| B. nealsonii | Nicotiana attenuate [27] |
| B. safensis | Osmanthus fragrans [28], Musa sp. [29], P. cuspidatum [19], S. officinarum [20], Chloris virgata [30] and V. radiata [26] |
| B. simplex | P. cuspidatum [19] |
| B. taxi | Taxus chinensis [31] |
| B. toyonensis | S. lycopersicum [18] |
| Paenibacillus etheri | - |
| P. humicus | Acacia sp. [32] and Eucalyptus spp. [24] |
| P. marcantiophytorum | Herbertus sendtneri [33] |
| P. pabuli | P. cuspidatum [19], S. officinarum [20] and V. radiata [26] |
| P. taichungensis | V. radiata [26] |
| P. poae | Vitis vinifera [34] |
| Sphingomonas. panni | Musa sp. [29] |
| Staphylococcus capitis subsp. capitis | G. hirsutum and Z. mays [25] |
| S. epidermidis | S. lycopersicum [35] and Musa sp. [29] |
| P. avellanae | Corylus avellane [36,37] |
| Isolate | Siderophores Production | Phosphate Solubilization | Ammonia Production | IAA Production | IAA µg mL−1 |
|---|---|---|---|---|---|
| Au01 (Pseudomonas avellanae) | + | - | 2 | + | 10.98 ± 2.44 |
| Au02 (Bacillus toyonensis) | - | - | 2 | - | 2.57 ± 1.81 |
| Au03 (Paenibacillus humicus) | - | - | 0 | - | 1.90 ± 1.81 |
| Au04 (Bacillus megaterium) | - | + | 2 | + | 6.22 ± 4.69 |
| Au05 (Bacillus toyonensis) | - | - | 2 | - | 3.10 ± 0.83 |
| Au06 (Bacillus cereus) | + | - | 2 | - | 1.72 ± 1.75 |
| Au08 (Bacillus toyonensis) | - | - | 2 | - | 4.40 ± 5.30 |
| Au14 (Bacillus safensis) | + | - | 2 | - | 2.21 ± 0.59 |
| Au15 (Paenibacillus pabuli) | - | - | 0 | - | 3.07 ± 1.42 |
| Au21 (Bacillus simplex) | - | - | 2 | + | 5.21 ± 4.36 |
| Au32 (Paenibacillus marchantiophytorum) | - | + | 1 | - | 0.56 ± 0.43 |
| Au39 (Pseudomonas poae) | + | + | 2 | + | 4.68 ± 2.11 |
| Au43 (Sphingomonas panni) | + | - | 1 | + | 6.17 ± 4.34 |
| Au47 (Paenibacillus etheri) | - | - | 0 | - | 0.74 ± 0.03 |
| Au53 (Paenibacillus taichungensis) | - | - | 0 | - | 2.47 ± 2.33 |
| Au61 (Bacillus taxi) | - | + | 0 | + | 3.87 ± 1.76 |
| Control (Pseudomonas syringae) | + | - | 2 | + | 8.43 ± 1.11 |
| Control (Escherichia coli) | + | 2 | - | 0 | |
| Negative Control | - | - | 0 | - | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martins, J.; Ares, A.; Casais, V.; Costa, J.; Canhoto, J. Identification and Characterization of Arbutus unedo L. Endophytic Bacteria Isolated from Wild and Cultivated Trees for the Biological Control of Phytophthora cinnamomi. Plants 2021, 10, 1569. https://doi.org/10.3390/plants10081569
Martins J, Ares A, Casais V, Costa J, Canhoto J. Identification and Characterization of Arbutus unedo L. Endophytic Bacteria Isolated from Wild and Cultivated Trees for the Biological Control of Phytophthora cinnamomi. Plants. 2021; 10(8):1569. https://doi.org/10.3390/plants10081569
Chicago/Turabian StyleMartins, João, Aitana Ares, Vinicius Casais, Joana Costa, and Jorge Canhoto. 2021. "Identification and Characterization of Arbutus unedo L. Endophytic Bacteria Isolated from Wild and Cultivated Trees for the Biological Control of Phytophthora cinnamomi" Plants 10, no. 8: 1569. https://doi.org/10.3390/plants10081569
APA StyleMartins, J., Ares, A., Casais, V., Costa, J., & Canhoto, J. (2021). Identification and Characterization of Arbutus unedo L. Endophytic Bacteria Isolated from Wild and Cultivated Trees for the Biological Control of Phytophthora cinnamomi. Plants, 10(8), 1569. https://doi.org/10.3390/plants10081569