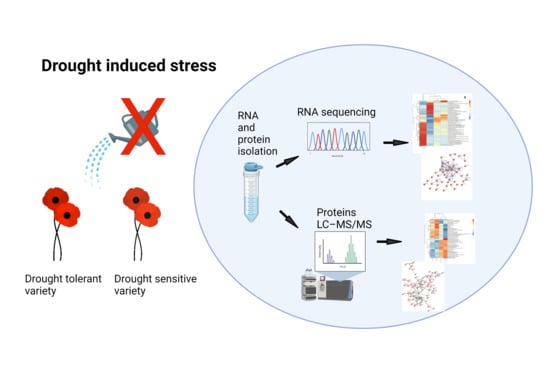
Transcriptomic and Proteomic Analysis of Drought Stress Response in Opium Poppy Plants during the First Week of Germination
Abstract
:1. Introduction
2. Results
2.1. Varieties of Papaver Somniferum
2.2. Gene and Protein Expression of Dehydrins
2.3. Identification of DEGs and DEPs
2.4. Interaction Networks and Biosynthetic Pathways within DEGs and DEPs
3. Discussion
4. Materials and Methods
4.1. Plant Material and Stress Treatments
4.2. RNA Extraction and DEGs Screening
4.3. Protein Isolation and DEPs Screening
4.4. Protein Functional Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; Von Korff, M. Leaf Proteome Alterations in the Context of Physiological and Morphological Responses to Drought and Heat Stress in Barley (Hordeum vulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef] [Green Version]
- Zougmoré, R.B.; Partey, S.T.; Ouédraogo, M.; Torquebiau, E.; Campbell, B.M. Facing Climate Variability in Sub-Saharan Africa: Analysis of Climate-Smart Agriculture Opportunities to Manage Climate-Related Risks. Cah. Agric. 2018, 27, 3400. [Google Scholar] [CrossRef] [Green Version]
- Neha, P.; Kumar, M.; Solankey, S.S. Impact of Drought and Salinity on Vegetable Crops and Mitigation Strategies. In Advances in Research on Vegetable Production Under a Changing Climate Vol. 1; Solankey, S.S., Kumari, M., Kumar, M., Eds.; Advances in Olericulture; Springer International Publishing: Cham, Switzerland, 2021; pp. 235–253. ISBN 978-3-030-63497-1. [Google Scholar]
- Singh, D.; Laxmi, A. Transcriptional Regulation of Drought Response: A Tortuous Network of Transcriptional Factors. Front. Plant Sci. 2015, 6, 895. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Q.; Cao, C.; Shen, T.; Zhong, L.; He, H.; Chen, X. Comprehensive Metabolomic and Proteomic Analysis in Biochemical Metabolic Pathways of Rice Spikes under Drought and Submergence Stress. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2019, 1867, 237–247. [Google Scholar] [CrossRef]
- Dias, M.C.; Pinto, D.C.G.A.; Figueiredo, C.; Santos, C.; Silva, A.M.S. Phenolic and Lipophilic Metabolite Adjustments in Olea Europaea (Olive) Trees during Drought Stress and Recovery. Phytochemistry 2021, 185, 112695. [Google Scholar] [CrossRef]
- Fahad, S.; Bajwa, A.A.; Nazir, U.; Anjum, S.A.; Farooq, A.; Zohaib, A.; Sadia, S.; Nasim, W.; Adkins, S.; Saud, S. Crop Production under Drought and Heat Stress: Plant Responses and Management Options. Front. Plant Sci. 2017, 8, 1147. [Google Scholar] [CrossRef] [Green Version]
- Anjum, S.A.; Ashraf, U.; Tanveer, M.; Khan, I.; Hussain, S.; Shahzad, B.; Zohaib, A.; Abbas, F.; Saleem, M.F.; Ali, I. Drought Induced Changes in Growth, Osmolyte Accumulation and Antioxidant Metabolism of Three Maize Hybrids. Front. Plant Sci. 2017, 8, 69. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B. Reactive Species and Antioxidants. Redox Biology Is a Fundamental Theme of Aerobic Life. Plant Physiol. 2006, 141, 312–322. [Google Scholar] [CrossRef] [Green Version]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmood, T.; Khalid, S.; Abdullah, M.; Ahmed, Z.; Shah, M.K.N.; Ghafoor, A.; Du, X. Insights into Drought Stress Signaling in Plants and the Molecular Genetic Basis of Cotton Drought Tolerance. Cells 2020, 9, 105. [Google Scholar] [CrossRef] [Green Version]
- Pour-Benab, S.M.; Fabriki-Ourang, S.; Mehrabi, A.-A. Expression of Dehydrin and Antioxidant Genes and Enzymatic Antioxidant Defense under Drought Stress in Wild Relatives of Wheat. Biotechnol. Biotechnol. Equip. 2019, 33, 1063–1073. [Google Scholar] [CrossRef] [Green Version]
- Ennajeh, M.; Vadel, A.M.; Khemira, H. Osmoregulation and Osmoprotection in the Leaf Cells of Two Olive Cultivars Subjected to Severe Water Deficit. Acta Physiol. Plant. 2009, 31, 711–721. [Google Scholar] [CrossRef]
- Cuevas-Velazquez, C.L.; Reyes, J.L.; Covarrubias, A.A. Group 4 Late Embryogenesis Abundant Proteins as a Model to Study Intrinsically Disordered Proteins in Plants. Plant Signal. Behav. 2017, 12, e1343777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodziewicz, P.; Swarcewicz, B.; Chmielewska, K.; Wojakowska, A.; Stobiecki, M. Influence of Abiotic Stresses on Plant Proteome and Metabolome Changes. Acta Physiol. Plant. 2014, 36, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Yang, Y.; Liu, D.; Wang, X.; Zhang, L. Transcription Factor TabHLH49 Positively Regulates Dehydrin WZY2 Gene Expression and Enhances Drought Stress Tolerance in Wheat. BMC Plant Biol. 2020, 20, 259. [Google Scholar] [CrossRef] [PubMed]
- Abedini, R.; GhaneGolmohammadi, F.; PishkamRad, R.; Pourabed, E.; Jafarnezhad, A.; Shobbar, Z.-S.; Shahbazi, M. Plant Dehydrins: Shedding Light on Structure and Expression Patterns of Dehydrin Gene Family in Barley. J. Plant Res. 2017, 130, 747–763. [Google Scholar] [CrossRef] [PubMed]
- Kumar, T.; Tiwari, N.; Bharadwaj, C.; Sarker, A.; Pappula, S.P.R.; Singh, S.; Singh, M. Identification of Allelic Variation in Drought Responsive Dehydrin Gene Based on Sequence Similarity in Chickpea (Cicer arietinum L.). Front. Genet. 2020, 11, 1579. [Google Scholar] [CrossRef]
- Čepl, J.; Stejskal, J.; Korecký, J.; Hejtmánek, J.; Faltinová, Z.; Lstibůrek, M.; Gezan, S. The Dehydrins Gene Expression Differs across Ecotypes in Norway Spruce and Relates to Weather Fluctuations. Sci. Rep. 2020, 10, 20789. [Google Scholar] [CrossRef]
- Vaseva, I.; Akiscan, Y.; Demirevska, K.; Anders, I.; Feller, U. Drought Stress Tolerance of Red and White Clover–Comparative Analysis of Some Chaperonins and Dehydrins. Sci. Hortic. 2011, 130, 653–659. [Google Scholar] [CrossRef]
- Beaudoin, G.A.; Facchini, P.J. Benzylisoquinoline Alkaloid Biosynthesis in Opium Poppy. Planta 2014, 240, 19–32. [Google Scholar] [CrossRef] [Green Version]
- Marciano, M.A.; Panicker, S.X.; Liddil, G.D.; Lindgren, D.; Sweder, K.S. Development of a Method to Extract Opium Poppy (Papaver somniferum L.) DNA from Heroin. Sci. Rep. 2018, 8, 2509. [Google Scholar]
- Zhang, Z.; Li, C.; Zhang, J.; Chen, F.; Gong, Y.; Li, Y.; Su, Y.; Wei, Y.; Zhao, Y. Selection of the Reference Gene for Expression Normalization in Papaver Somniferum L. under Abiotic Stress and Hormone Treatment. Genes 2020, 11, 124. [Google Scholar] [CrossRef] [Green Version]
- Dąbrowski, G.; Czaplicki, S.; Konopka, I. Composition and Quality of Poppy (Papaver somniferum L.) Seed Oil Depending on the Extraction Method. LWT 2020, 134, 110167. [Google Scholar] [CrossRef]
- Solanki, G.; Dodiya, N.S.; Kunwar, R.; Bisen, P.; Kumar, R.; Singh, J.S. Character Association and Path Coefficient Analysis for Seed Yield and Latex Yield in Opium Poppy (Papaver somniferum L.). Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 1116–1123. [Google Scholar] [CrossRef]
- Özcan, M.M.; Atalay, Ç. Determination of Seed and Oil Properties of Some Poppy (Papaver somniferum L.) Varieties. Grasas Y Aceites 2006, 57, 169–174. [Google Scholar]
- Prochazka, P.; Smutka, L. Czech Republic as an Important Producer of Poppy Seed. Agris-Line Pap. Econ. Inform. 2012, 4, 35–47. [Google Scholar]
- Zhao, Y.; Gao, C.; Shi, F.; Yun, L.; Jia, Y.; Wen, J. Transcriptomic and Proteomic Analyses of Drought Responsive Genes and Proteins in Agropyron Mongolicum Keng. Curr. Plant Biol. 2018, 14, 19–29. [Google Scholar] [CrossRef]
- Li, C.; Wang, Z.; Nong, Q.; Lin, L.; Xie, J.; Mo, Z.; Huang, X.; Song, X.; Malviya, M.K.; Solanki, M.K.; et al. Physiological Changes and Transcriptome Profiling in Saccharum Spontaneum L. Leaf under Water Stress and Re-Watering Conditions. Sci. Rep. 2021, 11, 5525. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, M.; Li, F.; Lv, H.; Li, C.; Xia, G. A Proteomic Study of the Response to Salinity and Drought Stress in an Introgression Strain of Bread Wheat. Mol. Cell. Proteom. 2009, 8, 2676–2686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wang, Y.; Wang, L.; Zhu, J.; Deng, J.; Tang, R.; Chen, G. Integration of Transcriptomic and Proteomic Analyses for Finger Millet [Eleusine Coracana (L.) Gaertn.] in Response to Drought Stress. PLoS ONE 2021, 16, e0247181. [Google Scholar] [CrossRef]
- Andersen, S.B. Plant Breeding from Laboratories to Fields; University of Copenhagen: Copenhagen, Denmark, 2013; ISBN 978-953-51-1090-3. [Google Scholar]
- Riyazuddin, R.; Nisha, N.; Singh, K.; Verma, R.; Gupta, R. Involvement of Dehydrin Proteins in Mitigating the Negative Effects of Drought Stress in Plants. Plant Cell Rep. 2021. [Google Scholar] [CrossRef]
- Davik, J.; Koehler, G.; From, B.; Torp, T.; Rohloff, J.; Eidem, P.; Wilson, R.C.; Sønsteby, A.; Randall, S.K.; Alsheikh, M. Dehydrin, Alcohol Dehydrogenase, and Central Metabolite Levels Are Associated with Cold Tolerance in Diploid Strawberry (Fragaria Spp.). Planta 2013, 237, 265–277. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Nam, K.H. Physiological Roles of ERD10 in Abiotic Stresses and Seed Germination of Arabidopsis. Plant Cell Rep. 2010, 29, 203–209. [Google Scholar] [CrossRef]
- Šunderlíková, V.; Wilhelm, E. High Accumulation of Legumin and Lea-like MRNAs during Maturation Is Associated with Increased Conversion Frequency of Somatic Embryos from Pedunculate Oak (Quercus robur L.). Protoplasma 2002, 220, 97–103. [Google Scholar] [CrossRef]
- Wang, R.Y.; Wang, H.G.; Liu, X.Y.; Lian, S.; Chen, L.; Qiao, Z.J.; McInerney, C.E.; Wang, L. Drought-Induced Transcription of Resistant and Sensitive Common Millet Varieties. J. Anim. Plant Sci. 2017, 27, 1303–1314. [Google Scholar]
- Morales-Merida, B.E.; Villicaña, C.; Perales-Torres, A.L.; Martínez-Montoya, H.; Castillo-Ruiz, O.; León-Chan, R.; Lighbourn-Rojas, L.A.; Heredia, J.B.; León-Félix, J. Transcriptomic Analysis in Response to Combined Stress by UV-B Radiation and Cold in Belle Pepper (Capsicum annuum). Int. J. Agric. Biol. 2021, 25, 1–12. [Google Scholar]
- Bettencourt, B.R.; Hogan, C.C.; Nimali, M.; Drohan, B.W. Inducible and Constitutive Heat Shock Gene Expression Responds to Modification of Hsp70 Copy Number in Drosophila Melanogaster but Does Not Compensate for Loss of Thermotolerance in Hsp70 Null Flies. BMC Biol. 2008, 6, 5. [Google Scholar] [CrossRef] [Green Version]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A Graphical Gene-Set Enrichment Tool for Animals and Plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Department of Primary Industries, Parks, Water and Environment. Best Practice Poppy Growing Guide. 2019. Available online: https://dpipwe.tas.gov.au/agriculture/plant-industries/best-practice-poppy-growing-guide (accessed on 10 August 2021).
- Verma, G.; Srivastava, D.; Tiwari, P.; Chakrabarty, D. ROS Modulation in Crop Plants Under Drought Stress. In Reactive Oxygen, Nitrogen and Sulfur Species in Plants; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 311–336. ISBN 978-1-119-46867-7. [Google Scholar]
- Deeba, F.; Pandey, A.K.; Ranjan, S.; Mishra, A.; Singh, R.; Sharma, Y.K.; Shirke, P.A.; Pandey, V. Physiological and Proteomic Responses of Cotton (Gossypium herbaceum L.) to Drought Stress. Plant Physiol. Biochem. 2012, 53, 6–18. [Google Scholar] [CrossRef]
- Sharma, S.; Villamor, J.G.; Verslues, P.E. Essential Role of Tissue-Specific Proline Synthesis and Catabolism in Growth and Redox Balance at Low Water Potential. Plant Physiol. 2011, 157, 292–304. [Google Scholar] [CrossRef] [Green Version]
- Blum, A. Osmotic Adjustment Is a Prime Drought Stress Adaptive Engine in Support of Plant Production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef]
- Graether, S.P.; Boddington, K.F. Disorder and Function: A Review of the Dehydrin Protein Family. Front. Plant Sci. 2014, 5, 576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, M.; Shinoda, Y.; Tanaka, Y.; Kuboi, T. DNA Binding of Citrus Dehydrin Promoted by Zinc Ion. Plant Cell Environ. 2009, 32, 532–541. [Google Scholar] [CrossRef] [PubMed]
- Saucedo, A.L.; Hernández-Domínguez, E.E.; de Luna-Valdez, L.A.; Guevara-García, A.A.; Escobedo-Moratilla, A.; Bojorquéz-Velázquez, E.; Del Río-Portilla, F.; Fernández-Velasco, D.A.; Barba de la Rosa, A.P. Insights on Structure and Function of a Late Embryogenesis Abundant Protein from Amaranthus Cruentus: An Intrinsically Disordered Protein Involved in Protection against Desiccation, Oxidant Conditions, and Osmotic Stress. Front. Plant Sci. 2017, 8, 497. [Google Scholar] [CrossRef] [PubMed]
- Jing, H.; Li, C.; Ma, F.; Ma, J.-H.; Khan, A.; Wang, X.; Zhao, L.-Y.; Gong, Z.-H.; Chen, R.-G. Genome-Wide Identification, Expression Diversication of Dehydrin Gene Family and Characterization of CaDHN3 in Pepper (Capsicum annuum L.). PLoS ONE 2016, 11, e0161073. [Google Scholar] [CrossRef] [PubMed]
- Egerton-Warburton, L.M.; Balsamo, R.A.; Close, T.J. Temporal Accumulation and Ultrastructural Localization of Dehydrins in Zea Mays. Physiol. Plant. 1997, 101, 545–555. [Google Scholar] [CrossRef]
- Verma, G.; Dhar, Y.V.; Srivastava, D.; Kidwai, M.; Chauhan, P.S.; Bag, S.K.; Asif, M.H.; Chakrabarty, D. Genome-Wide Analysis of Rice Dehydrin Gene Family: Its Evolutionary Conservedness and Expression Pattern in Response to PEG Induced Dehydration Stress. PLoS ONE 2017, 12, e0176399. [Google Scholar] [CrossRef] [Green Version]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic Acid Signaling Pathway in Plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Wang, R.; Liu, W.; Zhang, H.; Guo, Y.; Wen, R. Genome-Wide Characterization of Heat-Shock Protein 70s from Chenopodium Quinoa and Expression Analyses of Cqhsp70s in Response to Drought Stress. Genes 2018, 9, 35. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Yokoya, S. Enhanced Tolerance to Drought Stress in Transgenic Rice Plants Overexpressing a Small Heat-Shock Protein, SHSP17.7. Plant Cell Rep. 2008, 27, 329–334. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The Heat-Shock Protein/Chaperone Network and Multiple Stress Resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Ye, S.; Yu, S.; Shu, L.; Wu, J.; Wu, A.; Luo, L. Expression Profile Analysis of 9 Heat Shock Protein Genes throughout the Life Cycle and under Abiotic Stress in Rice. Chin. Sci. Bull. 2012, 57, 336–343. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Cai, G.; Kong, F.; Deng, Y.; Ma, N.; Meng, Q. Overexpression of Tomato Chloroplast-Targeted DnaJ Protein Enhances Tolerance to Drought Stress and Resistance to Pseudomonas Solanacearum in Transgenic Tobacco. Plant Physiol. Biochem. 2014, 82, 95–104. [Google Scholar] [CrossRef]
- Xia, Z.; Zhang, X.; Li, J.; Su, X.; Liu, J. Overexpression of a Tobacco J-Domain Protein Enhances Drought Tolerance in Transgenic Arabidopsis. Plant Physiol. Biochem. 2014, 83, 100–106. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K.; Shinozaki, K. The Transcriptional Regulatory Network in the Drought Response and Its Crosstalk in Abiotic Stress Responses Including Drought, Cold, and Heat. Front. Plant Sci. 2014, 5, 170. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cissé, N.; et al. Insight into the AP2/ERF Transcription Factor Superfamily in Sesame and Expression Profiling of DREB Subfamily under Drought Stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.; Zhang, B.; Qin, F. Arabidopsis RZFP34/CHYR1, a Ubiquitin E3 Ligase, Regulates Stomatal Movement and Drought Tolerance via SnRK2.6-Mediated Phosphorylation. Plant Cell 2015, 27, 3228–3244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, S.; Li, M.; Guan, Q.; Liu, F.; Zhang, S.; Chen, W.; Yin, L.; Qin, Y.; Ma, F. Physiological and Proteome Analysis Suggest Critical Roles for the Photosynthetic System for High Water-Use Efficiency under Drought Stress in Malus. Plant Sci. 2015, 236, 44–60. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, M.B.; Ahmad, Z.; Rashid, B.; Hassan, S.; Gregersen, P.L.; Leyva, M.D.; Nagy, I.; Asp, T.; Husnain, T. De Novo Assembly of Agave Sisalana Transcriptome in Response to Drought Stress Provides Insight into the Tolerance Mechanisms. Sci. Rep. 2019, 9, 396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The Flavonoid Biosynthetic Pathway in Arabidopsis: Structural and Genetic Diversity. Plant Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Halbwirth, H. The Creation and Physiological Relevance of Divergent Hydroxylation Patterns in the Flavonoid Pathway. Int. J. Mol. Sci. 2010, 11, 595–621. [Google Scholar] [CrossRef]
- Yao, X.; Wang, T.; Wang, H.; Liu, H.; Liu, S.; Zhao, Q.; Chen, K.; Zhang, P. Identification, Characterization and Expression Analysis of the Chalcone Synthase Family in the Antarctic Moss Pohlia nutans. Antarct. Sci. 2019, 31, 23–33. [Google Scholar] [CrossRef]
- Jiang, L.; Gao, Q.C.; Chen, Z.P.; Zhang, J.J.; Bai, X.Y.; He, X.L.; Xu, Q.X. Selenium Tolerance of an Arabidopsis Drought-Resistant Mutant Csm1-1. Russ. J. Plant Physiol. 2015, 62, 625–631. [Google Scholar] [CrossRef]
- Azzouz-Olden, F.; Hunt, A.G.; Dinkins, R. Transcriptome Analysis of Drought-Tolerant Sorghum Genotype SC56 in Response to Water Stress Reveals an Oxidative Stress Defense Strategy. Mol. Biol. Rep. 2020, 47, 3291–3303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salekdeh, G.H.; Siopongco, J.; Wade, L.J.; Ghareyazie, B.; Bennett, J. Proteomic Analysis of Rice Leaves during Drought Stress and Recovery. Proteomics 2002, 2, 1131–1145. [Google Scholar] [CrossRef]
- Shiriga, K.; Sharma, R.; Chaudhary, K.; Yadav, S.; Hossain, F. Expression Pattern of Superoxide Dismutase Under Drought Stress in Maize. Int. J. Innov. Res. Sci. Eng. Technol. 2014, 3, 11333–11337. [Google Scholar]
- Weng, M.; Cui, L.; Liu, F.; Zhang, M.; Shan, L.; Yang, S.; Deng, X.-P. Effects of Drought Stress on Antioxidant Enzymes in Seedlings of Different Wheat Genotypes. Pak. J. Bot. 2015, 47, 49–56. [Google Scholar]
- Bolwell, G.P.; Bozak, K.; Zimmerlin, A. Plant Cytochrome P450. Phytochemistry 1994, 37, 1491–1506. [Google Scholar] [CrossRef]
- Ghodke, P.; Khandagale, K.; Thangasamy, A.; Kulkarni, A.; Narwade, N.; Shirsat, D.; Randive, P.; Roylawar, P.; Singh, I.; Gawande, S.J.; et al. Comparative Transcriptome Analyses in Contrasting Onion (Allium cepa L.) Genotypes for Drought Stress. PLoS ONE 2020, 15, e0237457. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Ding, J.; Lee, D.; Lu, X.; Feng, Y.; Song, W. Overexpression of SoCYP85A1, a Spinach Cytochrome P450 Gene in Transgenic Tobacco Enhances Root Development and Drought Stress Tolerance. Front. Plant Sci. 2017, 8, 1909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greenbaum, D.; Colangelo, C.; Williams, K.; Gerstein, M. Comparing Protein Abundance and MRNA Expression Levels on a Genomic Scale. Genome Biol. 2003, 4, 117. [Google Scholar] [CrossRef] [Green Version]
- Koussounadis, A.; Langdon, S.P.; Um, I.H.; Harrison, D.J.; Smith, V.A. Relationship between Differentially Expressed MRNA and MRNA-Protein Correlations in a Xenograft Model System. Sci. Rep. 2015, 5, 10775. [Google Scholar] [CrossRef] [Green Version]
- de Sousa Abreu, R.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global Signatures of Protein and MRNA Expression Levels. Mol. Biosyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef] [Green Version]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Araújo, M.; Prada, J.; Mariz-Ponte, N.; Santos, C.; Pereira, J.A.; Pinto, D.C.G.A.; Silva, A.M.S.; Dias, M.C. Antioxidant Adjustments of Olive Trees (Olea europaea) under Field Stress Conditions. Plants 2021, 10, 684. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING V10: Protein–Protein Interaction Networks, Integrated over the Tree of Life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]




| Name of DHN | Accession Number | log2 Fold Change | AdjPVal | log2 Fold Change | AdjPVal | Stress Response According to Literature |
|---|---|---|---|---|---|---|
| dehydrin COR47-like | XP_026394044.1 | 0.9643 | 0.033 | −0.1393 | 0.0089 | Cold response [34] |
| dehydrin ERD10-like | XP_026397211.1 | 1.0916 | 0.0284 | −0.1368 | 0.0008 | Cold response, dehydration [35] |
| dehydrin DHN1-like | XP_026393601.1 | −0.0677 | 0.9843 | −0.2548 | 0.0005 | Drought stress [36] |
| dehydrin DHN1-like | XP_026432270.1 | 2.294 | 0.2181 | −0.1929 | 0.0028 | Drought stress [37] |
| dehydrin HIRD11-like | XP_026417500.1 | −0.2862 | 0.7536 | - | - | Cold response, UV-B radiation [38] |
| dehydrin HIRD11-like | XP_026426999.1 | −3.4628 | 0.6364 | - | - | Cold response, UV-B radiation [38] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kundrátová, K.; Bartas, M.; Pečinka, P.; Hejna, O.; Rychlá, A.; Čurn, V.; Červeň, J. Transcriptomic and Proteomic Analysis of Drought Stress Response in Opium Poppy Plants during the First Week of Germination. Plants 2021, 10, 1878. https://doi.org/10.3390/plants10091878
Kundrátová K, Bartas M, Pečinka P, Hejna O, Rychlá A, Čurn V, Červeň J. Transcriptomic and Proteomic Analysis of Drought Stress Response in Opium Poppy Plants during the First Week of Germination. Plants. 2021; 10(9):1878. https://doi.org/10.3390/plants10091878
Chicago/Turabian StyleKundrátová, Kristýna, Martin Bartas, Petr Pečinka, Ondřej Hejna, Andrea Rychlá, Vladislav Čurn, and Jiří Červeň. 2021. "Transcriptomic and Proteomic Analysis of Drought Stress Response in Opium Poppy Plants during the First Week of Germination" Plants 10, no. 9: 1878. https://doi.org/10.3390/plants10091878
APA StyleKundrátová, K., Bartas, M., Pečinka, P., Hejna, O., Rychlá, A., Čurn, V., & Červeň, J. (2021). Transcriptomic and Proteomic Analysis of Drought Stress Response in Opium Poppy Plants during the First Week of Germination. Plants, 10(9), 1878. https://doi.org/10.3390/plants10091878