Proline Affects Flowering Time in Arabidopsis by Modulating FLC Expression: A Clue of Epigenetic Regulation?
Abstract
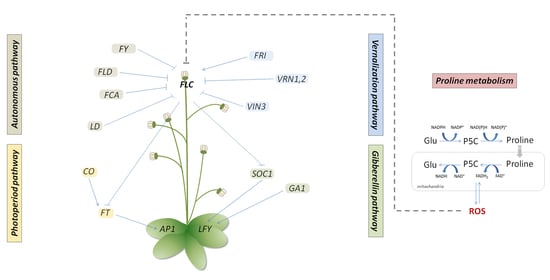
:1. Introduction
2. Results
2.1. FLC Is Upregulated in the Proline Biosynthesis Mutant p5cs1 p5cs2/P5CS2
2.2. Proline-Deficient Mutants Exhibit a Trichome Distribution Reminiscent of Plants Overexpressing FLC
2.3. Vernalization Experiments Show That FLC Is Necessary for Proline-Mediated Flowering
2.4. Flc-7 p5cs1 p5cs2/P5CS2 Mutants Flower as Early as the Flc-7 Parental Mutant
3. Discussion
4. Materials and Methods
4.1. Plant Growth Conditions, Physiological Experiments, and Plant Treatments
4.2. Molecular Techniques and Real-Time RT-qPCR
4.3. Identification and Characterization of Flc-7 T-DNA Insertion Mutant
4.4. Plant Genetic Crosses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bauduin, S.; Latini, M.; Belleggia, I.; Migliore, M.; Biancucci, M.; Mattioli, R.; Francioso, A.; Mosca, L.; Funck, D.; Trovato, M. Interplay between Proline Metabolism and ROS in the Fine Tuning of Root-Meristem Size in Arabidopsis. Plants 2022, 11, 1512. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Marchese, D.; D’Angeli, S.; Altamura, M.M.; Costantino, P.; Trovato, M. Modulation of intracellular proline levels affects flowering time and inflorescence architecture in Arabidopsis. Plant Mol. Biol. 2008, 66, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Székely, G.; Ábrahám, E.; Cséplő, A.; Rigó, G.; Zsigmond, L.; Csiszár, J.; Ayaydin, F.; Strizhov, N.; Jásik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J. 2008, 53, 11–28. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Falasca, G.; Sabatini, S.; Altamura, M.M.; Costantino, P.; Trovato, M. The proline biosynthetic genes P5CS1 and P5CS2 play overlapping roles in Arabidopsis flower transition but not in embryo development. Physiol. Plant 2009, 137, 72–85. [Google Scholar] [CrossRef] [PubMed]
- Funck, D.; Winter, G.; Baumgarten, L.; Forlani, G. Requirement of proline synthesis during Arabidopsis reproductive development. BMC Plant Biol. 2012, 12, 191. [Google Scholar] [CrossRef]
- Mattioli, R.; Biancucci, M.; Lonoce, C.; Costantino, P.; Trovato, M. Proline is required for male gametophyte development in Arabidopsis. BMC Plant Biol 2012, 12, 236. [Google Scholar] [CrossRef]
- Mattioli, R.; Palombi, N.; Funck, D.; Trovato, M. Proline Accumulation in Pollen Grains as Potential Target for Improved Yield Stability Under Salt Stress. Front. Plant Sci. 2020, 11, 582877. [Google Scholar] [CrossRef]
- Considine, M.J.; Foyer, C.H. Redox regulation of plant development. Antioxid. Redox Signal. 2014, 21, 1305–1326. [Google Scholar] [CrossRef]
- Gapper, C.; Dolan, L. Control of Plant Development by Reactive Oxygen Species. Plant Physiol. 2006, 141, 341–345. [Google Scholar] [CrossRef]
- Decros, G.; Baldet, P.; Beauvoit, B.; Stevens, R.; Flandin, A.; Colombié, S.; Gibon, Y.; Pétriacq, P. Get the Balance Right: ROS Homeostasis and Redox Signalling in Fruit. Front. Plant Sci. 2019, 10, 1091. [Google Scholar] [CrossRef] [Green Version]
- Dunand, C.; Crèvecoeur, M.; Penel, C. Distribution of superoxide and hydrogen peroxide in Arabidopsis root and their influence on root development: Possible interaction with peroxidases. New Phytol. 2007, 174, 332–341. [Google Scholar] [CrossRef] [PubMed]
- Tsukagoshi, H.; Busch, W.; Benfey, P.N. Transcriptional Regulation of ROS Controls Transition from Proliferation to Differentiation in the Root. Cell 2010, 143, 606–616. [Google Scholar] [CrossRef]
- Yamada, M.; Han, X.; Benfey, P.N. RGF1 controls root meristem size through ROS signalling. Nature 2020, 577, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Pasternak, T.; Eiblmeier, M.; Ditengou, F.; Kochersperger, P.; Sun, J.; Wang, H.; Rennenberg, H.; Teale, W.; Paponov, I.; et al. Plastid-localized glutathione reductase2-regulated glutathione redox status is essential for Arabidopsis root apical meristem maintenance. Plant Cell 2013, 25, 4451–4468. [Google Scholar] [CrossRef] [PubMed]
- Nakanomyo, I.; Kost, B.; Chua, N.-H.; Fukuda, H. Preferential and Asymmetrical Accumulation of a Rac Small GTPase mRNA in Differentiating Xylem Cells of Zinnia elegans. Plant Cell Physiol. 2002, 43, 1484–1492. [Google Scholar] [CrossRef]
- Dennery, P.A. Effects of oxidative stress on embryonic development. Birth Defects Res. Part C Embryo Today Rev. 2007, 81, 155–162. [Google Scholar] [CrossRef]
- Bailly, C. The signalling role of ROS in the regulation of seed germination and dormancy. Biochem. J. 2019, 476, 3019–3032. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Ju, Y.; Kessler, S.A. Reactive Oxygen Species as Mediators of Gametophyte Development and Double Fertilization in Flowering Plants. Front. Plant Sci. 2020, 11, 1199. [Google Scholar] [CrossRef]
- Xie, D.-L.; Zheng, X.-L.; Zhou, C.-Y.; Kanwar, M.K.; Zhou, J. Functions of Redox Signaling in Pollen Development and Stress Response. Antioxidants 2022, 11, 287. [Google Scholar] [CrossRef]
- Guan, C.; Huang, Y.H.; Cui, X.; Liu, S.J.; Zhou, Y.Z.; Zhang, Y.W. Overexpression of gene encoding the key enzyme involved in proline-biosynthesis (PuP5CS) to improve salt tolerance in switchgrass (Panicum virgatum L.). Plant Cell Rep. 2018, 37, 1187–1199. [Google Scholar] [CrossRef]
- Simpson, G.G.; Dean, C. Arabidopsis, the Rosetta Stone of Flowering Time? Science 2002, 296, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Boss, P.K.; Bastow, R.M.; Mylne, J.S.; Dean, C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 2004, 16 (Suppl. S1), S18–S31. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; Amasino, R.M. Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 2004, 427, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Bäurle, I.; Dean, C. The Timing of Developmental Transitions in Plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Leijten, W.; Koes, R.; Roobeek, I.; Frugis, G. Translating Flowering Time from Arabidopsis thaliana to Brassicaceae and Asteraceae Crop Species. Plants 2018, 7, 111. [Google Scholar] [CrossRef] [PubMed]
- Pajoro, A.; Biewers, S.; Dougali, E.; Leal Valentim, F.; Mendes, M.A.; Porri, A.; Coupland, G.; Van de Peer, Y.; van Dijk, A.D.; Colombo, L.; et al. The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: A two-decade history. J. Exp. Bot. 2014, 65, 4731–4745. [Google Scholar] [CrossRef]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef]
- Poethig, R.S. Phase change and the regulation of shoot morphogenesis in plants. Science 1990, 250, 923–930. [Google Scholar] [CrossRef]
- Bloomer, R.H.; Dean, C. Fine-tuning timing: Natural variation informs the mechanistic basis of the switch to flowering in Arabidopsis thaliana. J. Exp. Bot. 2017, 68, 5439–5452. [Google Scholar] [CrossRef]
- Chiang, H.H.; Dandekar, A.M. Regulation of proline accumulation in Arabidopsis thaliana (L.) Heynh during development and in response to desiccation. Plant Cell Environ. 1995, 18, 1280–1290. [Google Scholar] [CrossRef]
- Schwacke, R.; Grallath, S.; Breitkreuz, K.E.; Stransky, E.; Stransky, H.; Frommer, W.B.; Rentsch, D. LeProT1, a transporter for proline, glycine betaine, and gamma-amino butyric acid in tomato pollen. Plant Cell 1999, 11, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Nanjo, T.; Kobayashi, M.; Yoshiba, Y.; Sanada, Y.; Wada, K.; Tukaya, H.; Kakubari, Y.; Yamaguchi- Shinozaki, K.; Shinozaki, K. Biological functions of proline in morphogenesis and osmotolerance revealed in antisense transgenic Arabidopsis thaliana. Plant J. 1999, 18, 185–193. [Google Scholar] [CrossRef] [PubMed]
- Samach, A.; Onouchi, H.; Gold, S.E.; Ditta, G.S.; Schwarz-Sommer, Z.S.; Yanofsky, M.F.; Coupland, G. Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 2000, 288, 1613–1616. [Google Scholar] [CrossRef] [PubMed]
- Zdunek-Zastocka, E.; Grabowska, A.; Branicki, T.; Michniewska, B. Biochemical characterization of the triticale TsPAP1, a new type of plant prolyl aminopeptidase, and its impact on proline content and flowering time in transgenic Arabidopsis plants. Plant Physiol. Biochem. 2017, 116, 18–26. [Google Scholar] [CrossRef]
- Bastow, R.; Mylne, J.S.; Lister, C.; Lippman, Z.; Martienssen, R.A.; Dean, C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 2004, 427, 164–167. [Google Scholar] [CrossRef]
- Ramakrishnan, M.; Papolu, P.K.; Satish, L.; Vinod, K.K.; Wei, Q.; Sharma, A.; Emamverdian, A.; Zou, L.-H.; Zhou, M. Redox status of the plant cell determines epigenetic modifications under abiotic stress conditions and during developmental processes. J. Adv. Res. 2022. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 15 June 2022).
- Ahmed, M.; Kim, D.R. pcr: An R package for quality assessment, analysis and testing of qPCR data. PeerJ. 2018, 6, e4473. [Google Scholar] [CrossRef]
- Willmann, M.R.; Poethig, R.S. The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development 2011, 138, 677–685. [Google Scholar] [CrossRef]
- Michaels, S.D.; Amasino, R.M. FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 1999, 11, 949–956. [Google Scholar] [CrossRef]
- Helliwell, C.A.; Wood, C.C.; Robertson, M.; James Peacock, W.; Dennis, E.S. The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J. 2006, 46, 183–192. [Google Scholar] [CrossRef]
- Hepworth, S.R.; Valverde, F.; Ravenscroft, D.; Mouradov, A.; Coupland, G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002, 21, 4327–4337. [Google Scholar] [CrossRef] [PubMed]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Kröber, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed]
- Kleinboelting, N.; Huep, G.; Kloetgen, A.; Viehoever, P.; Weisshaar, B. GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012, 40, D1211–D1215. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Christensen, H.E.; Ishimaru, Y.; Dong, C.H.; Chao-Ming, W.; Cleary, A.L.; Chua, N.H. Profilin plays a role in cell elongation, cell shape maintenance, and flowering in Arabidopsis. Plant Physiol. 2000, 124, 1637–1647. [Google Scholar] [CrossRef]
- Pandey, D.K.; Chaudhary, B. Domestication-driven Gossypium profilin 1 (GhPRF1) gene transduces early flowering phenotype in tobacco by spatial alteration of apical/floral-meristem related gene expression. BMC Plant Biol. 2016, 16, 112. [Google Scholar] [CrossRef]
- Dietzel, L.; Gläßer, C.; Liebers, M.; Hiekel, S.; Courtois, F.; Czarnecki, O.; Schlicke, H.; Zubo, Y.; Börner, T.; Mayer, K.; et al. Identification of Early Nuclear Target Genes of Plastidial Redox Signals that Trigger the Long-Term Response of Arabidopsis to Light Quality Shifts. Mol. Plant 2015, 8, 1237–1252. [Google Scholar] [CrossRef]
- Rai, K.K.; Rai, N.; Rai, S.P. Response of Lablab purpureus L. to high temperature stress and role of exogenous protectants in mitigating high temperature induced oxidative damages. Mol. Biol. Rep. 2018, 45, 1375–1395. [Google Scholar] [CrossRef]
- Sabbioni, G.; Funck, D.; Forlani, G. Enzymology and Regulation of δ1-Pyrroline-5-Carboxylate Synthetase 2 From Rice. Front. Plant Sci. 2021, 12, 672702. [Google Scholar] [CrossRef]
- Donald, S.P.; Sun, X.Y.; Hu, C.A.; Yu, J.; Mei, J.M.; Valle, D.; Phang, J.M. Proline oxidase, encoded by p53-induced gene-6, catalyzes the generation of proline-dependent reactive oxygen species. Cancer Res. 2001, 61, 1810–1815. [Google Scholar]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The Plant NADPH Oxidase RBOHD Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef]
- Hare, P.; Cress, W. Metabolic implications of stress-induced proline accumulation in plants. Plant Growth Regul. 1997, 21, 79–102. [Google Scholar] [CrossRef]
- Shetty, K.; Wahlqvist, M.L. A model for the role of the proline-linked pentose-phosphate pathway in phenolic phytochemical bio-synthesis and mechanism of action for human health and environmental applications. Asia Pac. J. Clin. Nutr. 2004, 13, 1–24. [Google Scholar] [PubMed]
- Rédei, G.P. Supervital Mutants of Arabidopsis. Genetics 1962, 47, 443–460. [Google Scholar] [CrossRef] [PubMed]
- Koornneef, M.; Hanhart, C.J.; van der Veen, J.H. A genetic and physiological analysis of late flowering mutants in Arabidopsis thaliana. Mol. Gen. Genet. 1991, 229, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Stewart, C.N.; Via, L.E. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques 1993, 14, 748–750. [Google Scholar]





| Full Gene Name | Acronym | AGI Locus Code | Floral Pathway |
|---|---|---|---|
| GIGANTEA | GI | AT1G22770 | Photoperiodic |
| FLOWERING WAGENINGEN | FWA | AT4G25530 | Photoperiodic |
| CONSTANS | CO | AT5G15840 | Photoperiodic |
| LUMINIDEPENDENS | LD | AT4G02560 | Autonomous |
| FLOWERING CONTROL LOCUS A | FCA | AT4G16280 | Autonomous |
| FLOWERING LOCUS KH DOMAIN | FLK | AT3G04610 | Autonomous |
| FLOWERING LOCUS D | FLD | AT3G10390 | Autonomous |
| Unknown | FPA | AT2G43410 | Autonomous |
| Unknown | FVE | AT2G19520 | Autonomous |
| Unknown | FY | AT5G13480 | Autonomous |
| REDUCED VERNALIZATION RESPONSE 1 | VRN1 | AT3G18990 | Vernalization |
| REDUCED VERNALIZATION RESPONSE 2 | VRN2 | AT4G16845 | Vernalization |
| VERNALIZATION INSENSITIVE 3 | VIN3 | AT5G57380 | Vernalization |
| GA REQUIRING 1 | GA1 | AT4G02780 | GA-dependent |
| GIBBERELLIN 3-OXIDASE 1 | GA3ox1 | AT1G15550 | GA-dependent |
| GIBBERELLIN 3-OXIDASE 2 | GA3ox2 | AT1G80340 | GA-dependent |
| GIBBERELLIN 3-OXIDASE 3 | GA3ox3 | AT4G21690 | GA-dependent |
| GIBBERELLIN 3-OXIDASE 4 | GA3ox4 | AT1G80330 | GA-dependent |
| GIBBERELLIN 20 OXIDASE 1 | GA20ox1 | AT4G25420 | GA-dependent |
| GIBBERELLIN 20 OXIDASE 2 | GA20ox2 | AT5G51810 | GA-dependent |
| GIBBERELLIN 2-OXIDASE 1 | GA2ox1 | AT1G78440 | GA-dependent |
| FLOWERING LOCUS T | FT | AT1G65480 | Floral integrator |
| FLOWERING LOCUS C | FLC | AT5G10140 | Floral integrator |
| SUPPRESSOR OF OVEREXPRESSION OF CO1 | SOC1 | AT2G45660 | Floral integrator |
| LEAFY | LFY | AT5G61850 | Floral integrator |
| APETALA 1 | AP1 | AT1G26310 | Meristem identity |
| CAULIFLOWER | CAL | AT1G26310 | Meristem identity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mattioli, R.; Francioso, A.; Trovato, M. Proline Affects Flowering Time in Arabidopsis by Modulating FLC Expression: A Clue of Epigenetic Regulation? Plants 2022, 11, 2348. https://doi.org/10.3390/plants11182348
Mattioli R, Francioso A, Trovato M. Proline Affects Flowering Time in Arabidopsis by Modulating FLC Expression: A Clue of Epigenetic Regulation? Plants. 2022; 11(18):2348. https://doi.org/10.3390/plants11182348
Chicago/Turabian StyleMattioli, Roberto, Antonio Francioso, and Maurizio Trovato. 2022. "Proline Affects Flowering Time in Arabidopsis by Modulating FLC Expression: A Clue of Epigenetic Regulation?" Plants 11, no. 18: 2348. https://doi.org/10.3390/plants11182348
APA StyleMattioli, R., Francioso, A., & Trovato, M. (2022). Proline Affects Flowering Time in Arabidopsis by Modulating FLC Expression: A Clue of Epigenetic Regulation? Plants, 11(18), 2348. https://doi.org/10.3390/plants11182348