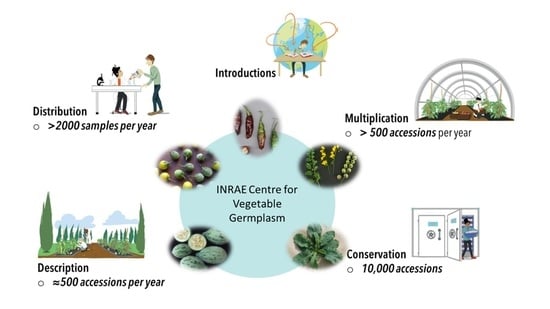
The INRAE Centre for Vegetable Germplasm: Geographically and Phenotypically Diverse Collections and Their Use in Genetics and Plant Breeding
Abstract
:1. Introduction
2. Overview and Origins of the Five Collections
3. Summary of the Individual Collections
- (i).
- The aubergine collection (2388 accessions) contains two introductory peaks: one from 1991, which corresponds to the scientific collaboration with the taxonomist Richard N. Lester of the University of Birmingham, UK, and the second in 2004, when the collection of Solanum species related to aubergine were transferred from the University of Birmingham to INRAE [17]. More recently, we have introduced new aubergine accessions from the H2020 G2P-SOL project (http://www.g2p-sol.eu/ (accessed on 5 December 2021)). Most of the aubergine accessions originate from Africa and Asia. The aubergine collection is notable amongst the five collections because it includes a large proportion of crop wild relatives (26%). This number of CWR is rare in other germplasm centres, mostly because of the difficulties in maintaining them. The collection includes more than 1000 accessions principally from Africa (related cultivated species including Solanum macrocarpon L., Solanum aethiopicum L., Solanum scabrum Mill.). These species are indigenous African leafy vegetables and/or fruits: S. aethiopicum and S. macrocarpon provide a usable secondary gene pool for the improvement of S. melongena. Approximately 500 accessions, representing over a hundred wild species, the majority related to cultivated aubergine, complete the collection, as well as accessions of other Solanaceae of interest (Atropa, Datura, Lycium, Nicandra, Physalis and Withania) [18]. A word of caution must be added about the taxonomy of Solanum species. The taxonomic classification of aubergine dates back to the work of RN Lester in the 1990s; the nomenclature has evolved since the early 2000s and the taxonomic status of several taxa is unclear [19,20].
- (ii).
- The pepper collection (2188 accessions) is representative of domestication centres (South and Central America). In pepper, the collection focuses on Capsicum annuum (76% of accessions), with a large collection, rich in phenotypic and geographic variability, that is easily exploited in breeding programmes. Eleven species of Capsicum are available in the collection, including the five cultivated species (C. annuum, C. frutescens, C. chinense, C. baccatum and C. pubescens) [21]. Recombinant inbred lines in pepper [22] have allowed the evaluation of fruit traits [23] and resistance to Phytophthora species [24,25]. Similarly to aubergine, 912 INRAE accessions (889 cultivated) were included in the G2P-SOL project, of which 59 are included in the final core collection [26]. The genotypic diversity in pepper from the G2P-SOL core collection maximises the diversity of around 10,000 accessions from 10 genebanks and research institutes from around the world in a collection of 423 mostly C. annuum accessions [26]. INRAE is the official distributor of the G2P-SOL pepper core collection. Another core collection of over 280 accessions has been constructed with INRAE material [27].
- (iii).
- The tomato collection (3410 accessions) is representative of its domestication centre (South and Central America). In tomato, the number of wild-relative species is lower than in the other collections but a good diversity of S. pimpinellifolium is available. For tomato, S. peruvianum has been separated into four species, including two new species, S. arcanum and S. huaylasense, which requires database information to be corrected: taxonomic identification is therefore an ongoing process [28]. More than 500 accessions have been genotyped with the SolCap Illumina array and a core collection of 160 accessions constructed and amply characterised (see below) [29]. The tomato scientific resources include progenies of recombinant inbred lines [30], advanced backcrosses, intra- and interspecific progenies and multi-parent progenies (MAGIC) [31] representing more than 1000 accessions.
- (iv).
- The melon collection (2359 accessions) comes from all around the world, particularly Africa and Asia, and includes around 100 genotypes of wild C. melo (mostly from the agrestis cultigroup), which are compatible for crossing with cultivated melons. Recombinant inbred lines obtained by crossing distant melon lines have been created and studied for many segregating agronomic traits and for monogenic as well as quantitative pest and disease resistance [32,33,34,35,36,37]. A mutant melon collection obtained by chemical EMS mutagenesis of an INRAE Charentais melon line includes more than 7000 M2 families and is useful for the functional validation of genes or for generating new diversity [38,39,40,41].
- (v).
- The lettuce collection (948 accessions) comes essentially from Europe. For cultivated lettuce, the introduction year is unknown for 151 cultivars received before 1980; for wild Lactuca species, 64% of the wild accessions (mainly of L. serriola) were collected directly by INRAE, mainly in France in the 1980s. Out of the 479 introduced wild accessions, 343 collected accessions and 136 received from other laboratories, only 248 are still present with seed stock in the 2020s: seed for many accessions arriving in the 1970s was lost because the storage at room temperature before 1984 was inadequate for long-term conservation in Lactuca. In lettuce, the 704 cultivated accessions are L. sativa, with many modern cultivars cultivated in Western Europe over the last 40 years. For wild lettuce, the precise collection site is known for 92% of the INRAE accessions. There are 11 Lactuca species, including the three species mainly used by breeders: L. serriola, L. saligna and L. virosa. The lettuce collection is completed by 15 accessions from other genera of the Asteraceae family (Chondrilla, Mycelis, Sonchus). The lettuce collection contains a few lines with resistance to potyviruses or Bremia lactuca identified in L. virosa and introgressed into a cultivated background [42,43,44].
4. Collection Management
- (i).
- Conservation of seed stocks
- (ii).
- Regeneration of seed stocks
- (iii).
- Descriptions
- (iv).
- Networks and sub-collections
- (v).
- Seed and data sharing
5. The Collections as Material for Scientific Study
- (i).
- Domestication and structure of the collections
- (ii).
- Resistance to plant pests and pathogens
- (iii).
- Floral biology and crossing compatibility
- (iv).
- Fruit quality and abiotic stress tolerance
- (v).
- Selection and breeding
6. Conclusions and Perspectives
- -
- Allows the study of allelic diversity for genes of interest for mining allelic diversity;
- -
- Facilitates the determination of the “uniqueness” of an accession and better identification of duplicates;
- -
- Gives an idea of the phenomenon of introgression between species;
- -
- Can help in the study of core- and pan-genomes.
- -
- Increasing the duration between multiplication cycles by improved conservation;
- -
- Mobile applications for management of collections (seed harvest, descriptions in the field, etc.) and their direct link to databases for improved traceability and quality standards with the aim of obtaining ISO 9001 certification;
- -
- More complete and quantifiable phenotyping by use of image analysis to measure size, shape and colour of plant organs.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Aubriot, X.; Knapp, S.; Syfert, M.M.; Poczai, P.; Buerki, S. Shedding New Light on the Origin and Spread of the Brinjal Eggplant (Solanum melongena L.) and Its Wild Relatives. Am. J. Bot. 2018, 105, 1175–1187. [Google Scholar] [CrossRef] [Green Version]
- Kraft, K.H.; Brown, C.H.; Nabhan, G.P.; Luedeling, E.; De Luna Ruiz, J.J.; Coppens d’Eeckenbrugge, G.; Hijmans, R.J.; Gepts, P. Multiple Lines of Evidence for the Origin of Domesticated Chili Pepper, Capsicum annuum, in Mexico. Proc. Natl. Acad. Sci. USA 2014, 111, 6165–6170. [Google Scholar] [CrossRef] [Green Version]
- Razifard, H.; Ramos, A.; Della Valle, A.L.; Bodary, C.; Goetz, E.; Manser, E.J.; Li, X.; Zhang, L.; Visa, S.; Tieman, D.; et al. Genomic Evidence for Complex Domestication History of the Cultivated Tomato in Latin America. Mol. Biol. Evol. 2020, 37, 1118–1132. [Google Scholar] [CrossRef]
- Endl, J.; Achigan-Dako, E.G.; Pandey, A.K.; Monforte, A.J.; Pico, B.; Schaefer, H. Repeated Domestication of Melon (Cucumis melo) in Africa and Asia and a New Close Relative from India. Am. J. Bot. 2018, 105, 1662–1671. [Google Scholar] [CrossRef] [Green Version]
- Zhao, G.; Lian, Q.; Zhang, Z.; Fu, Q.; He, Y.; Ma, S.; Ruggieri, V.; Monforte, A.; Wang, P.; Julca, I.; et al. A Comprehensive Genome Variation Map of Melon Identifies Multiple Domestication Events and Loci Influencing Agronomic Traits. Nat. Genet. 2019, 51, 1607–1615. [Google Scholar] [CrossRef]
- Wei, T.; Van Treuren, R.; Liu, X.; Zhang, Z.; Chen, J.; Liu, Y.; Dong, S.; Sun, P.; Yang, T.; Lan, T.; et al. Whole-Genome Resequencing of 445 Lactuca Accessions Reveals the Domestication History of Cultivated Lettuce. Nat. Genet. 2021, 53, 752–760. [Google Scholar] [CrossRef]
- De Vries, I.M. Origin and Domestication of Lactuca sativa L. Genet. Resour. Crop. Evol. 1997, 44, 165–174. [Google Scholar] [CrossRef]
- Lebeda, A.; Ryder, E.J.; Grube, R.; Dolezalova, I.; Kristova, E. Lettuce (Asteraceae; Lactuca Spp.). In Genetic Resources, Chromosome Engineering, and Crop Improvement. Vegetable Crops; Genetic Resources, Chromosome Engineering, and Crop Improvement Series; CRC Press: Boca Raton, FL, USA; Taylor & Francis Group: Oxford, UK, 2007; pp. 377–472. ISBN 978-0-8493-9646-5. [Google Scholar]
- Mou, B. Lettuce. Vegetables 1: “Asteraceae”, “Brassicaceae”, “Chenopodicaceae”, and “Cucurbitaceae”. In Handbook of Plant Breeding; Springer: New York, NY, USA, 2008; pp. 73–114. ISBN 978-0-387-30443-4. [Google Scholar]
- Paran, I.; Van der Knaap, E. Genetic and Molecular Regulation of Fruit and Plant Domestication Traits in Tomato and Pepper. J. Exp. Bot. 2007, 58, 3841–3852. [Google Scholar] [CrossRef] [Green Version]
- Herms, D.A.; Mattson, W.J. The Dilemma of Plants: To Grow or Defend. Q. Rev. Biol. 1992, 67, 283–335. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Lindhout, P. Domestication and Breeding of Tomatoes: What Have We Gained and What Can We Gain in the Future? Ann. Bot. 2007, 100, 1085–1094. [Google Scholar] [CrossRef]
- Biosafety Unit About the Nagoya Protocol. Available online: https://www.cbd.int/abs/about/ (accessed on 5 December 2021).
- Tontisirin, K.; Nantel, G.; Bhattacharjee, L. Food-Based Strategies to Meet the Challenges of Micronutrient Malnutrition in the Developing World. Proc. Nutr. Soc. 2002, 61, 243–250. [Google Scholar] [CrossRef]
- Quenouille, J.; Saint-Felix, L.; Moury, B.; Palloix, A. Diversity of Genetic Backgrounds Modulating the Durability of a Major Resistance Gene. Analysis of a Core Collection of Pepper Landraces Resistant to Potato Virus Y. Mol. Plant Pathol. 2016, 17, 296–302. [Google Scholar] [CrossRef]
- Sauvage, C.; Segura, V.; Bauchet, G.; Stevens, R.; Do, P.T.; Nikoloski, Z.; Fernie, A.R.; Causse, M. Genome-Wide Association in Tomato Reveals 44 Candidate Loci for Fruit Metabolic Traits. Plant Physiol. 2014, 165, 1120–1132. [Google Scholar] [CrossRef] [Green Version]
- Lester, R.N.; Hawkes, J.G.; Marsh, S.; Daunay, M.C.; Van der Weerden, G.; Barendse, G. The Sources, Successes and Successors of the Birmingham University Solanaceae Collection (1964–2000). In Solanaceae V, Advances in Taxonomy and Utilization; Nijmegen University Press: Nijmegen, The Netherlands, 2001; pp. 391–412. ISBN 978-90-373-0580-7. [Google Scholar]
- Salinier, J.; Brand-Daunay, M.-C.; Goussopoulos, J.; Gros, C.; Lefebvre, V. The Fascinating Diversity of Pepper and Eggplant Collections Held at the INRA Centre for Vegetable Germplasm. In Proceedings of the Eucarpia Meeting on Genetics and Breeding of Capsicum and Eggplant, Avignon, France, 11–13 September 2019. [Google Scholar]
- Daunay, M.C.; Lester, R.N.; Van der Weerden, G. La taxinomie des Solanacées. Ce qu’il faut savoir et les pièges à éviter. In Virus des Solanacées: Du Génome Viral à la Protection des Cultures; Synthèses; Éd. Quae: Versailles, France, 2008; pp. 3–10. ISBN 978-2-7592-0076-4. [Google Scholar]
- Aubriot, X.; Daunay, M.-C. Eggplants and Relatives: From Exploring Their Diversity and Phylogenetic Relationships to Conservation Challenges. In The Eggplant Genome; Compendium of Plant Genomes; Chapman, M.A., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2019; pp. 91–134. ISBN 978-3-319-99207-5. [Google Scholar]
- Nicolaï, M.; Cantet, M.; Lefebvre, V.; Sage-Palloix, A.-M.; Palloix, A. Genotyping a Large Collection of Pepper (Capsicum Spp.) with SSR Loci Brings New Evidence for the Wild Origin of Cultivated C. Annuum and the Structuring of Genetic Diversity by Human Selection of Cultivar Types. Genet. Resour. Crop. Evol. 2013, 60, 2375–2390. [Google Scholar] [CrossRef]
- Barchi, L.; Bonnet, J.; Boudet, C.; Signoret, P.; Nagy, I.; Lanteri, S.; Palloix, A.; Lefebvre, V. A High-Resolution, Intraspecific Linkage Map of Pepper (Capsicum annuum L.) and Selection of Reduced Recombinant Inbred Line Subsets for Fast Mapping. Genome 2007, 50, 51–60. [Google Scholar] [CrossRef]
- Barchi, L.; Lefebvre, V.; Sage-Palloix, A.-M.; Lanteri, S.; Palloix, A. QTL Analysis of Plant Development and Fruit Traits in Pepper and Performance of Selective Phenotyping. Theor. Appl. Genet. 2009, 118, 1157–1171. [Google Scholar] [CrossRef]
- Bonnet, J.; Danan, S.; Boudet, C.; Barchi, L.; Sage-Palloix, A.-M.; Caromel, B.; Palloix, A.; Lefebvre, V. Are the Polygenic Architectures of Resistance to Phytophthora capsici and P. parasitica Independent in Pepper? Theor. Appl. Genet. 2007, 115, 253–264. [Google Scholar] [CrossRef]
- Thabuis, A.; Palloix, A.; Pflieger, S.; Daubèze, A.-M.; Caranta, C.; Lefebvre, V. Comparative Mapping of Phytophthora Resistance Loci in Pepper Germplasm: Evidence for Conserved Resistance Loci across Solanaceae and for a Large Genetic Diversity. Theor. Appl. Genet. 2003, 106, 1473–1485. [Google Scholar] [CrossRef]
- Tripodi, P.; Rabanus-Wallace, M.T.; Barchi, L.; Kale, S.; Esposito, S.; Acquadro, A.; Schafleitner, R.; Van Zonneveld, M.; Prohens, J.; Diez, M.J.; et al. Global Range Expansion History of Pepper (Capsicum Spp.) Revealed by over 10,000 Genebank Accessions. Proc. Natl. Acad. Sci. USA 2021, 118, e2104315118. [Google Scholar] [CrossRef]
- Tamisier, L.; Szadkowski, M.; Nemouchi, G.; Lefebvre, V.; Szadkowski, E.; Duboscq, R.; Santoni, S.; Sarah, G.; Sauvage, C.; Palloix, A.; et al. Genome-Wide Association Mapping of QTLs Implied in Potato Virus Y Population Sizes in Pepper: Evidence for Widespread Resistance QTL Pyramiding. Mol. Plant Pathol. 2020, 21, 3–16. [Google Scholar] [CrossRef]
- Peralta, I.E.; Knapp, S.; Spooner, D.M. New Species of Wild Tomatoes (Solanum lycopersicon: Solanaceae) from Northern Peru. Syst. Bot. 2005, 30, 424–434. [Google Scholar] [CrossRef]
- Sim, S.-C.; Durstewitz, G.; Plieske, J.; Wieseke, R.; Ganal, M.W.; Van Deynze, A.; Hamilton, J.P.; Buell, C.R.; Causse, M.; Wijeratne, S.; et al. Development of a Large SNP Genotyping Array and Generation of High-Density Genetic Maps in Tomato. PLoS ONE 2012, 7, e40563. [Google Scholar] [CrossRef] [PubMed]
- Saliba-Colombani, V.; Causse, M.; Langlois, D.; Philouze, J.; Buret, M. Genetic Analysis of Organoleptic Quality in Fresh Market Tomato. 1. Mapping QTLs for Physical and Chemical Traits. Theor. Appl. Genet. 2001, 102, 259–272. [Google Scholar] [CrossRef]
- Pascual, L.; Desplat, N.; Huang, B.E.; Desgroux, A.; Bruguier, L.; Bouchet, J.-P.; Le, Q.H.; Chauchard, B.; Verschave, P.; Causse, M. Potential of a Tomato MAGIC Population to Decipher the Genetic Control of Quantitative Traits and Detect Causal Variants in the Resequencing Era. Plant Biotechnol. J. 2015, 13, 565–577. [Google Scholar] [CrossRef]
- Boissot, N.; Thomas, S.; Sauvion, N.; Marchal, C.; Pavis, C.; Dogimont, C. Mapping and Validation of QTLs for Resistance to Aphids and Whiteflies in Melon. Theor. Appl. Genet. 2010, 121, 9–20. [Google Scholar] [CrossRef]
- Diaz, A.; Fergany, M.; Formisano, G.; Ziarsolo, P.; Blanca, J.; Fei, Z.; Staub, J.E.; Zalapa, J.E.; Cuevas, H.E.; Dace, G.; et al. A Consensus Linkage Map for Molecular Markers and Quantitative Trait Loci Associated with Economically Important Traits in Melon (Cucumis melo L.). BMC Plant Biol. 2011, 11, 111. [Google Scholar] [CrossRef] [Green Version]
- Perchepied, L.; Dogimont, C.; Pitrat, M. Strain-Specific and Recessive QTLs Involved in the Control of Partial Resistance to Fusarium oxysporum f. Sp. Melonis Race 1.2 in a Recombinant Inbred Line Population of Melon. Theor. Appl. Genet. 2005, 111, 65–74. [Google Scholar] [CrossRef]
- Périn, C.; Hagen, S.; De Conto, V.; Katzir, N.; Danin-Poleg, Y.; Portnoy, V.; Baudracco-Arnas, S.; Chadoeuf, J.; Dogimont, C.; Pitrat, M. A Reference Map of Cucumis melo Based on Two Recombinant Inbred Line Populations. Theor. Appl. Genet. 2002, 104, 1017–1034. [Google Scholar] [CrossRef]
- Périn, C.; Hagen, L.S.; Giovinazzo, N.; Besombes, D.; Dogimont, C.; Pitrat, M. Genetic Control of Fruit Shape Acts Prior to Anthesis in Melon (Cucumis melo L.). Mol. Genet. Genom. 2002, 266, 933–941. [Google Scholar] [CrossRef]
- Périn, C.; Gomez-Jimenez, M.; Hagen, L.; Dogimont, C.; Pech, J.-C.; Latché, A.; Pitrat, M.; Lelièvre, J.-M. Molecular and Genetic Characterization of a Non-Climacteric Phenotype in Melon Reveals Two Loci Conferring Altered Ethylene Response in Fruit. Plant Physiol. 2002, 129, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Boualem, A.; Fergany, M.; Fernandez, R.; Troadec, C.; Martin, A.; Morin, H.; Sari, M.-A.; Collin, F.; Flowers, J.M.; Pitrat, M.; et al. A Conserved Mutation in an Ethylene Biosynthesis Enzyme Leads to Andromonoecy in Melons. Science 2008, 321, 836–838. [Google Scholar] [CrossRef]
- Boualem, A.; Lemhemdi, A.; Sari, M.-A.; Pignoly, S.; Troadec, C.; Abou Choucha, F.; Solmaz, I.; Sari, N.; Dogimont, C.; Bendahmane, A. The Andromonoecious Sex Determination Gene Predates the Separation of Cucumis and Citrullus Genera. PLoS ONE 2016, 11, e0155444. [Google Scholar] [CrossRef] [Green Version]
- Dahmani-Mardas, F.; Troadec, C.; Boualem, A.; Lévêque, S.; Alsadon, A.A.; Aldoss, A.A.; Dogimont, C.; Bendahmane, A. Engineering Melon Plants with Improved Fruit Shelf Life Using the TILLING Approach. PLoS ONE 2010, 5, e15776. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Troadec, C.; Boualem, A.; Rajab, M.; Fernandez, R.; Morin, H.; Pitrat, M.; Dogimont, C.; Bendahmane, A. A Transposon-Induced Epigenetic Change Leads to Sex Determination in Melon. Nature 2009, 461, 1135–1138. [Google Scholar] [CrossRef]
- Maisonneuve, B.B.; Bellec, Y.; Souche, S.; Lot, H. New Resistance against Downy Mildew and Lettuce Mosaic Potyvirus in Wild Lactuca Spp. In Proceedings of the Eucarpia Leafy Vegetables 99, European Association for Research on Plant Breeding, Olomouc, Czech Republic, 8–11 June 1999. [Google Scholar]
- Maisonneuve, B.; Bellec, Y.; Martin, E.; Lot, H.; Gognalon, P.; Moury, B. Lettuce Lines with Potyvirus Resistance: Differential Set for Study of Strains and New Genitors of Resistance. In Proceedings of the Abstracts Eucarpia Leafy Vegetables 2019, Olomouc, Czech Republic, 24–28 June 2019; pp. 86–87. [Google Scholar]
- Maisonneuve, B.; Pitrat, M.; Gognalons, P.; Moury, B. Growth Stage-Dependent Resistance to the Potyviruses Lettuce Italian Necrotic Virus and Lettuce Mosaic Virus Displayed by Lactuca Sativa Introgression Lines Carrying the Mo3 Locus from L. Virosa. Plant Pathol. 2018, 67, 2013–2018. [Google Scholar] [CrossRef]
- Plant Production and Protection Division. Genebank Standards for Plant Genetic Resources for Food and Agriculture; FAO: Rome, Italy, 2016; ISBN 978-92-5-109528-7. [Google Scholar]
- Nanduri, K.; Hanson, J.; Dulloo, M.; Ghosh, K.; Nowell, D.; Larinde, M. Handbooks for Genebanks No. 8 Seed Handling in Genebanks; Bioversity International: Rome, Italy, 2006; Available online: https://www.bioversityinternational.org/e-library/publications/detail/manual-of-seed-handling-in-genebanks/ (accessed on 5 December 2021).
- Daunay, M.C.; Dalmon, A.; Lester, R.N. Management of a Collection of Solanum Species for Eggplant (Solanum melongena) Breeding Purposes. In Solanaceae IV: Advances in Biology and Utilization; Royal Botanic Gardens: Kew, UK, 1999; pp. 369–383. ISBN 978-1-900347-90-7. [Google Scholar]
- Syfert, M.M.; Castañeda-Álvarez, N.P.; Khoury, C.K.; Särkinen, T.; Sosa, C.C.; Achicanoy, H.A.; Bernau, V.; Prohens, J.; Daunay, M.-C.; Knapp, S. Crop Wild Relatives of the Brinjal Eggplant (Solanum melongena): Poorly Represented in Genebanks and Many Species at Risk of Extinction. Am. J. Bot. 2016, 103, 635–651. [Google Scholar] [CrossRef] [Green Version]
- Causse, M.; Saliba-Colombani, V.; Lecomte, L.; Duffe, P.; Rousselle, P.; Buret, M. QTL Analysis of Fruit Quality in Fresh Market Tomato: A Few Chromosome Regions Control the Variation of Sensory and Instrumental Traits. J. Exp. Bot. 2002, 53, 2089–2098. [Google Scholar] [CrossRef]
- Lefebvre, V.; Kuntz, M.; Camara, B.; Palloix, A. The Capsanthin-Capsorubin Synthase Gene: A Candidate Gene for the y Locus Controlling the Red Fruit Colour in Pepper. Plant Mol. Biol. 1998, 36, 785–789. [Google Scholar] [CrossRef]
- Pitrat, M. Phenotypic Diversity in Wild and Cultivated Melons (Cucumis melo). Plant Biotechnol. 2013, 30, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Salinier, J.; Daunay, M.C.; Talmot, V.; Lecarpentier, C.; Pagès, L.; Bardel, A.; Fournier, C.; Torres, M.; Stevens, R. Root Architectural Trait Diversity in Aubergine (Solanum melongena L.) and Related Species and Correlations with Plant Biomass. Crop. Breed. Genet. Genom. 2019, 1, e190011. [Google Scholar] [CrossRef] [Green Version]
- Arnoux, S.; Fraïsse, C.; Sauvage, C. Genomic Inference of Complex Domestication Histories in Three Solanaceae Species. J. Evol. Biol. 2021, 34, 270–283. [Google Scholar] [CrossRef]
- Davidar, P.; Snow, A.A.; Rajkumar, M.; Pasquet, R.; Daunay, M.-C.; Mutegi, E. The Potential for Crop to Wild Hybridization in Eggplant (Solanum melongena; Solanaceae) in Southern India. Am. J. Bot. 2015, 102, 129–139. [Google Scholar] [CrossRef] [Green Version]
- Mutegi, E.; Snow, A.A.; Rajkumar, M.; Pasquet, R.; Ponniah, H.; Daunay, M.-C.; Davidar, P. Genetic Diversity and Population Structure of Wild/Weedy Eggplant (Solanum insanum, Solanaceae) in Southern India: Implications for Conservation. Am. J. Bot. 2015, 102, 140–148. [Google Scholar] [CrossRef]
- Page, A.; Gibson, J.; Meyer, R.S.; Chapman, M.A. Eggplant Domestication: Pervasive Gene Flow, Feralization, and Transcriptomic Divergence. Mol. Biol. Evol. 2019, 36, 1359–1372. [Google Scholar] [CrossRef]
- Chakrabarti, M.; Zhang, N.; Sauvage, C.; Munos, S.; Blanca, J.; Canizares, J.; Diez, M.J.; Schneider, R.; Mazourek, M.; McClead, J.; et al. A Cytochrome P450 Regulates a Domestication Trait in Cultivated Tomato. Proc. Natl. Acad. Sci. USA 2013, 110, 17125–17130. [Google Scholar] [CrossRef] [Green Version]
- Frary, A.; Nesbitt, T.C.; Grandillo, S.; Knaap, E.; Cong, B.; Liu, J.; Meller, J.; Elber, R.; Alpert, K.B.; Tanksley, S.D. Fw2.2: A Quantitative Trait Locus Key to the Evolution of Tomato Fruit Size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef] [Green Version]
- Sauvage, C.; Rau, A.; Aichholz, C.; Chadoeuf, J.; Sarah, G.; Ruiz, M.; Santoni, S.; Causse, M.; David, J.; Glémin, S. Domestication Rewired Gene Expression and Nucleotide Diversity Patterns in Tomato. Plant J. 2017, 91, 631–645. [Google Scholar] [CrossRef] [Green Version]
- Blanca, J.; Montero-Pau, J.; Sauvage, C.; Bauchet, G.; Illa, E.; Díez, M.J.; Francis, D.; Causse, M.; Van der Knaap, E.; Cañizares, J. Genomic Variation in Tomato, from Wild Ancestors to Contemporary Breeding Accessions. BMC Genom. 2015, 16, 257. [Google Scholar] [CrossRef] [Green Version]
- Bauchet, G.; Munos, S.; Sauvage, C.; Bonnet, J.; Grivet, L.; Causse, M. Genes Involved in Floral Meristem in Tomato Exhibit Drastically Reduced Genetic Diversity and Signature of Selection. BMC Plant Biol. 2014, 14, 279. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.; Zhu, G.; Zhang, J.; Xu, X.; Yu, Q.; Zheng, Z.; Zhang, Z.; Lun, Y.; Li, S.; Wang, X.; et al. Genomic Analyses Provide Insights into the History of Tomato Breeding. Nat. Genet. 2014, 46, 1220–1226. [Google Scholar] [CrossRef]
- Serres-Giardi, L.; Dogimont, C. How Microsatellite Diversity Helps to Understand the Domestication History of Melon. In Proceedings of the Xth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, Antalya, Turkey, 15–18 October 2012; pp. 254–263. [Google Scholar]
- Paris, H.S.; Janick, J.; Daunay, M.-C. Medieval Herbal Iconography and Lexicography of Cucumis (Cucumber and Melon, Cucurbitaceae) in the Occident, 1300–1458. Ann. Bot. 2011, 108, 471–484. [Google Scholar] [CrossRef] [Green Version]
- Dhillon, N.P.S.; Ranjana, R.; Singh, K.; Eduardo, I.; Monforte, A.J.; Pitrat, M.; Dhillon, N.K.; Singh, P.P. Diversity among Landraces of Indian Snapmelon (Cucumis melo Var. Momordica). Genet. Resour. Crop. Evol. 2007, 54, 1267–1283. [Google Scholar] [CrossRef]
- Fergany, M.; Kaur, B.; Monforte, A.J.; Pitrat, M.; Rys, C.; Lecoq, H.; Dhillon, N.P.S.; Dhaliwal, S.S. Variation in Melon (Cucumis melo) Landraces Adapted to the Humid Tropics of Southern India. Genet. Resour. Crop. Evol. 2011, 58, 225–243. [Google Scholar] [CrossRef] [Green Version]
- Huet, G. Breeding for Resistances to Ralstonia solanacearum. Front. Plant Sci. 2014, 5, 715. [Google Scholar] [CrossRef] [Green Version]
- Mansfield, J.; Genin, S.; Magori, S.; Citovsky, V.; Sriariyanum, M.; Ronald, P.; Dow, M.; Verdier, V.; Beer, S.V.; Machado, M.A.; et al. Top 10 Plant Pathogenic Bacteria in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 614–629. [Google Scholar] [CrossRef] [Green Version]
- Xue, H.; Lozano-Durán, R.; Macho, A.P. Insights into the Root Invasion by the Plant Pathogenic Bacterium Ralstonia solanacearum. Plants 2020, 9, 516. [Google Scholar] [CrossRef]
- Clarke, C.R.; Studholme, D.J.; Hayes, B.; Runde, B.; Weisberg, A.; Cai, R.; Wroblewski, T.; Daunay, M.-C.; Wicker, E.; Castillo, J.A.; et al. Genome-Enabled Phylogeographic Investigation of the Quarantine Pathogen Ralstonia solanacearum Race 3 Biovar 2 and Screening for Sources of Resistance Against Its Core Effectors. Phytopathology® 2015, 105, 597–607. [Google Scholar] [CrossRef] [Green Version]
- Lebeau, A.; Daunay, M.-C.; Frary, A.; Palloix, A.; Wang, J.-F.; Dintinger, J.; Chiroleu, F.; Wicker, E.; Prior, P. Bacterial Wilt Resistance in Tomato, Pepper, and Eggplant: Genetic Resources Respond to Diverse Strains in the Ralstonia solanacearum Species Complex. Phytopathology 2011, 101, 154–165. [Google Scholar] [CrossRef] [Green Version]
- Lebeau, A.; Gouy, M.; Daunay, M.C.; Wicker, E.; Chiroleu, F.; Prior, P.; Frary, A.; Dintinger, J. Genetic Mapping of a Major Dominant Gene for Resistance to Ralstonia solanacearum in Eggplant. Theor. Appl. Genet. 2013, 126, 143–158. [Google Scholar] [CrossRef]
- Pensec, F.; Lebeau, A.; Daunay, M.C.; Chiroleu, F.; Guidot, A.; Wicker, E. Towards the Identification of Type III Effectors Associated with Ralstonia Solanacearum Virulence on Tomato and Eggplant. Phytopathology® 2015, 105, 1529–1544. [Google Scholar] [CrossRef] [Green Version]
- Salgon, S.; Jourda, C.; Sauvage, C.; Daunay, M.-C.; Reynaud, B.; Wicker, E.; Dintinger, J. Eggplant Resistance to the Ralstonia solanacearum Species Complex Involves Both Broad-Spectrum and Strain-Specific Quantitative Trait Loci. Front. Plant Sci. 2017, 8, 828. [Google Scholar] [CrossRef] [Green Version]
- Salgon, S.; Raynal, M.; Lebon, S.; Baptiste, J.-M.; Daunay, M.-C.; Dintinger, J.; Jourda, C. Genotyping by Sequencing Highlights a Polygenic Resistance to Ralstonia pseudosolanacearum in Eggplant (Solanum melongena L.). IJMS 2018, 19, 357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivey, M.L.; Madrid, A.J.; Daunay, M.-C.; Shah, D. Evaluation of Tomato, Eggplant and Pepper Accessions for Resistance to Ralstonia solanacearum Species Complex (RSSC) Strains from Louisiana. Eur. J. Plant Pathol. 2021, 159, 279. [Google Scholar] [CrossRef]
- Cantet, M. Variation Phénotypique de La Résistance Quantitative à Phytophthora capsici Dans La Diversité Naturelle Du Piment, et Diversité Moléculaire et Profil d’évolution Du QTL Majeur Pc5.1. Ph.D. Thesis, Institut National d’Etudes Supérieures Agronomiques de Montpellier, Montpellier, France, 12 April 2013. [Google Scholar]
- Mallard, S.; Cantet, M.; Massire, A.; Bachellez, A.; Ewert, S.; Lefebvre, V. A Key QTL Cluster Is Conserved among Accessions and Exhibits Broad-Spectrum Resistance to Phytophthora capsici: A Valuable Locus for Pepper Breeding. Mol. Breed. 2013, 32, 349–364. [Google Scholar] [CrossRef]
- Lefebvre, V.; Daubèze, A.-M.; Van der Voort, J.R.; Peleman, J.; Bardin, M.; Palloix, A. QTLs for Resistance to Powdery Mildew in Pepper under Natural and Artificial Infections. Theor. Appl. Genet. 2003, 107, 661–666. [Google Scholar] [CrossRef]
- Moury, B.; Pflieger, S.; Blattes, A.; Lefebvre, V.; Palloix, A. A CAPS Marker to Assist Selection of Tomato Spotted Wilt Virus (TSWV) Resistance in Pepper. Genome 2000, 43, 137–142. [Google Scholar] [CrossRef]
- Lefebvre, V.; Palloix, A.; Caranta, C.; Pochard, E. Construction of an Intraspecific Integrated Linkage Map of Pepper Using Molecular Markers and Doubled-Haploid Progenies. Genome 1995, 38, 112–121. [Google Scholar] [CrossRef]
- Caranta, C.; Pflieger, S.; Lefebvre, V.; Daubèze, A.M.; Thabuis, A.; Palloix, A. QTLs Involved in the Restriction of Cucumber Mosaic Virus (CMV) Long-Distance Movement in Pepper. Theor. Appl. Genet. 2002, 104, 586–591. [Google Scholar] [CrossRef]
- Caranta, C.; Palloix, A.; Lefebvre, V.; Daubèze, A.M. QTLs for a Component of Partial Resistance to Cucumber Mosaic Virus in Pepper: Restriction of Virus Installation in Host-Cells. Theor. Appl. Genet. 1997, 94, 431–438. [Google Scholar] [CrossRef]
- Caranta, C.; Lefebvre, V.; Palloix, A. Polygenic Resistance of Pepper to Potyviruses Consists of a Combination of Isolate-Specific and Broad-Spectrum Quantitative Trait Loci. MPMI 1997, 10, 872–878. [Google Scholar] [CrossRef] [Green Version]
- Lefebvre, V.; Palloix, A. Both Epistatic and Additive Effects of QTLs Are Involved in Polygenic Induced Resistance to Disease: A Case Study, the Interaction Pepper—Phytophthora capsici Leonian. Theor. Appl. Genet. 1996, 93, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Caranta, C.; Dogimont, C. Plant Resistance to Viruses: Natural Resistance Associated with Recessive Genes. In Encyclopedia of Virology; Elsevier: Amsterdam, The Netherlands, 2008; pp. 177–186. ISBN 978-0-12-374410-4. [Google Scholar]
- Pitrat, M. Melon Genetic Resources: Phenotypic Diversity and Horticultural Taxonomy. In Genetics and Genomics of Cucurbitaceae; Plant Genetics and Genomics: Crops and Models; Springer International Publishing: Berlin/Heidelberg, Germany, 2016; Volume 20, 340p. [Google Scholar]
- Romay, G.; Pitrat, M.; Lecoq, H.; Wipf-Scheibel, C.; Millot, P.; Girardot, G.; Desbiez, C. Resistance Against Melon Chlorotic Mosaic Virus and Tomato Leaf Curl New Delhi Virus in Melon. Plant Dis. 2019, 103, 2913–2919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solmaz, I.; Sari, N.; Dogimont, C.; Pitrat, M. Evaluation of Turkish Melon Accessions for Resistance to Fusarium Wilt, Downy Mildew, Powdery Mildew, Cucumber mosaic virus and Zucchini yellow mosaic virus. Acta Hortic. 2016, 133–140. [Google Scholar] [CrossRef]
- Villeneuve, F.; Latour, F.; Théry, T.; Steinberg, C.; Edel-Hermann, V.; Pitrat, M.; Daunay, M.-C. The Control of Soil-Borne Vascular Diseases: Limits of Genetic Resistance of Cultivars and Rootstocks for Controlling Fusarium oxysporum F. Sp. Melonis (Melon) and Verticillium Sp. (Eggplant). Acta Hortic. 2014, 57–65. [Google Scholar] [CrossRef]
- Yousif, M.T.; Kheyr-Pour, A.; Gronenborn, B.; Pitrat, M.; Dogimont, C. Sources of Resistance to Watermelon Chlorotic Stunt Virus in Melon. Plant Breed. 2007, 126, 422–427. [Google Scholar] [CrossRef]
- Dogimont, C.; Boissot, N. Insect Resistance in Melon and Its Modification by Molecular Breeding. In Functional Genomics and Biotechnology in Solanaceae and Cucurbitaceae Crops; Springer: Berlin/Heidelberg, Germany, 2016; pp. 199–219. ISBN 978-3-662-48535-4. [Google Scholar]
- Dogimont, C.; Bendahmane, A.; Chovelon, V.; Boissot, N. Host Plant Resistance to Aphids in Cultivated Crops: Genetic and Molecular Bases, and Interactions with Aphid Populations. Comptes Rendus Biol. 2010, 333, 566–573. [Google Scholar] [CrossRef] [PubMed]
- Dogimont, C.; Chovelon, V.; Pauquet, J.; Boualem, A.; Bendahmane, A. The Vat Locus Encodes for a CC-NBS-LRR Protein That Confers Resistance to Aphis gossypii Infestation and A. Gossypii-Mediated Virus Resistance. Plant J. 2014, 80, 993–1004. [Google Scholar] [CrossRef]
- Thomas, S.; Dogimont, C.; Boissot, N. Association between Aphis gossypii Genotype and Phenotype on Melon Accessions. Arthropod-Plant Interact. 2012, 6, 93–101. [Google Scholar] [CrossRef]
- Boissot, N.; Schoeny, A.; Vanlerberghe-Masutti, F. Vat, an Amazing Gene Conferring Resistance to Aphids and Viruses They Carry: From Molecular Structure to Field Effects. Front. Plant Sci. 2016, 7, 1420. [Google Scholar] [CrossRef] [Green Version]
- Desbiez, C.; Schoeny, A.; Maisonneuve, B.; Berthier, K.; Bornard, I.; Chandeysson, C.; Fabre, F.; Girardot, G.; Gognalons, P.; Lecoq, H.; et al. Molecular and Biological Characterization of Two Potyviruses Infecting Lettuce in Southeastern France. Plant Pathol. 2017, 66, 970–979. [Google Scholar] [CrossRef]
- Parra, L.; Maisonneuve, B.; Lebeda, A.; Schut, J.; Christopoulou, M.; Jeuken, M.; McHale, L.; Truco, M.-J.; Crute, I.; Michelmore, R. Rationalization of Genes for Resistance to Bremia lactucae in Lettuce. Euphytica 2016, 210, 309–326. [Google Scholar] [CrossRef]
- Maisonneuve, B.; Chassagne, C.; Gautheron, E.; Glaux, C.; Morris, C.E. Screening of Genetic Resources for Resistance to Xanthomonas axonopodis Pv. Vitians. In Proceedings of the Eucarpia Leafy Vegetables Conference, Warwick, UK, 18–20 April 2007. [Google Scholar]
- Maisonneuve, B.; Jouan, C.; Gard, B.; Gimenez, S.; Goillon, C.; Jean, L.; Trottin, Y.; Védie, H.; Pitrat, M. LACTUMEL–Des Résistances Aux Nématodes à Galles Du Genre Meloidogyne Dans Le Genre Lactuca: Espoir Pour La Sélection de La Laitue. Innov. Agron. 2021, 84, 11–23. [Google Scholar] [CrossRef]
- Maisonneuve, B. Lactuca Virosa, a Source of Disease Resistance Genes for Lettuce Breeding: Results and Difficulties for Gene Introgression. In Proceedings of the EUCARPIA Meeting on Leafy Vegetables Genetics and Breeding, Noordwijkerhout, The Netherlands, 19–21 March 2003; pp. 61–67. [Google Scholar]
- Maisonneuve, B.; Bellec, Y.; Anderson, P.; Michelmore, R.W. Rapid Mapping of Two Genes for Resistance to Downy Mildew from Lactuca serriola to Existing Clusters of Resistance Genes. Theor. Appl. Genet. 1994, 89, 96–104. [Google Scholar] [CrossRef]
- Wood, K.J.; Nur, M.; Gil, J.; Fletcher, K.; Lakeman, K.; Gann, D.; Gothberg, A.; Khuu, T.; Kopetzky, J.; Naqvi, S.; et al. Effector Prediction and Characterization in the Oomycete Pathogen Bremia lactucae Reveal Host-Recognized WY Domain Proteins That Lack the Canonical RXLR Motif. PLoS Pathog. 2020, 16, e1009012. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, B.; Allen-Aubouard, C.; Pitrat, M. Effect of Plant Genotype on the Efficacy of Stimulators of Plant Defences in Two Horticultural Pathosystems. IOBC WPRS Bull. 2013, 89, 327–331. [Google Scholar]
- Sandoya, G.V.; Maisonneuve, B.; Truco, M.J.; Bull, C.T.; Simko, I.; Trent, M.; Hayes, R.J.; Michelmore, R.W. Genetic Analysis of Resistance to Bacterial Leaf Spot in the Heirloom Lettuce Cultivar Reine Des Glaces. Mol. Breed. 2019, 39, 160. [Google Scholar] [CrossRef]
- Maisonneuve, B.B.; Chupeau, M.C.; Bellec, Y.; Chupeau, Y.Y. Sexual and Somatic Hybridization in the Genus Lactuca. Euphytica 1995, 85, 281–285. [Google Scholar] [CrossRef]
- Albert, E.; Segura, V.; Gricourt, J.; Bonnefoi, J.; Derivot, L.; Causse, M. Association Mapping Reveals the Genetic Architecture of Tomato Response to Water Deficit: Focus on Major Fruit Quality Traits. J. Exp. Bot. 2016, 67, 6413–6430. [Google Scholar] [CrossRef]
- Bauchet, G.; Grenier, S.; Samson, N.; Segura, V.; Kende, A.; Beekwilder, J.; Cankar, K.; Gallois, J.; Gricourt, J.; Bonnet, J.; et al. Identification of Major Loci and Genomic Regions Controlling Acid and Volatile Content in Tomato Fruit: Implications for Flavor Improvement. New Phytol. 2017, 215, 624–641. [Google Scholar] [CrossRef] [Green Version]
- Bauchet, G.; Grenier, S.; Samson, N.; Bonnet, J.; Grivet, L.; Causse, M. Use of Modern Tomato Breeding Germplasm for Deciphering the Genetic Control of Agronomical Traits by Genome Wide Association Study. Theor. Appl. Genet. 2017, 130, 875–889. [Google Scholar] [CrossRef]
- Xu, J.; Ranc, N.; Muños, S.; Rolland, S.; Bouchet, J.-P.; Desplat, N.; Le Paslier, M.-C.; Liang, Y.; Brunel, D.; Causse, M. Phenotypic Diversity and Association Mapping for Fruit Quality Traits in Cultivated Tomato and Related Species. Theor. Appl. Genet. 2013, 126, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Duangjit, J.; Causse, M.; Sauvage, C. Efficiency of Genomic Selection for Tomato Fruit Quality. Mol. Breed. 2016, 36, 29. [Google Scholar] [CrossRef]
- Albert, E.; Gricourt, J.; Bertin, N.; Bonnefoi, J.; Pateyron, S.; Tamby, J.-P.; Bitton, F.; Causse, M. Genotype by Watering Regime Interaction in Cultivated Tomato: Lessons from Linkage Mapping and Gene Expression. Theor. Appl. Genet. 2016, 129, 395–418. [Google Scholar] [CrossRef] [PubMed]
- Bineau, E.; Diouf, I.; Carretero, Y.; Duboscq, R.; Bitton, F.; Djari, A.; Zouine, M.; Causse, M. Genetic Diversity of Tomato Response to Heat Stress at the QTL and Transcriptome Levels. Plant J. 2021, 107, 1213–1227. [Google Scholar] [CrossRef]
- Diouf, I.; Derivot, L.; Koussevitzky, S.; Carretero, Y.; Bitton, F.; Moreau, L.; Causse, M. Genetic Basis of Phenotypic Plasticity and Genotype × Environment Interactions in a Multi-Parental Tomato Population. J. Exp. Bot. 2020, 71, 5365–5376. [Google Scholar] [CrossRef]
- Daunay, M.C.; Lester, R.N.; Dalmon, A.; Ferri, M.; Kapilima, W.; Poveda-Aguilar, M.M.; Jullian, E. The Use of Wild Genetic Resources for Eggplant (Solanum melongena L.) Breeding. II Crossability and Fertility of Interspecific Hybrids. In Proceedings of the Xth Meeting on Genetics and Breeding of Capsicum and Eggplant, Avignon, France, 7–11 September 1998; INRA: Paris, France, 1998; pp. 19–24, ISBN 978-2-7380-0830-5. [Google Scholar]
- Daunay, M.C. Eggplant. In Vegetables II: Fabaceae, Liliaceae, Solanaceae, and Umbelliferae; Handbook of Plant Breeding; Springer: New York, NY, USA, 2008; pp. 163–220. ISBN 978-0-387-74108-6. [Google Scholar]
- Daunay, M.C.; Lester, R.N.; Laterrot, H. The Use of Wild Species for the Genetic Improvement of Brinjal Eggplant (Solanum melongena) and Tomato (Lycopersicon esculentum). In Solanaceae III: Taxonomy, Chemistry, Evolution; [Based on Papers Read at the Third International Solanaceae Congress, held in Bogotá, Columbia from July 25 to 30, 1988]; The Royal Botanic Gardens: Kew, UK, 1991; pp. 389–412. ISBN 978-0-947643-31-7. [Google Scholar]
- Daunay, M.C.; Lester, R.N.; Gebhardt, C.; Hennart, J.W.; Jahn, M.; Frary, A.; Doganlar, S. Genetic Resources of Eggplant (Solanum melongena L.) and Allied Species: A New Challenge for Molecular Geneticists and Eggplant Breeders. In Solanaceae V: Advances in Taxonomy and Utilization; Nijmegen University Press: Nijmegen, The Netherlands, 2001; pp. 251–274. ISBN 978-90-373-0580-7. [Google Scholar]
- Lanteri, S.; Rotino, G.L. Breakthroughs in the Genetics and Breeding of Capsicum and Eggplant: Proceedings of the XV EUCARPIA Meeting on Genetics and Breeding of Capsicum and Eggplant: 2–4 September 2013, Torino-Italy; FA, Università degli Studi di Torino: Torino, Italy, 2013; ISBN 978-88-97239-16-1. [Google Scholar]
- Villeneuve, F.; Latour, F.; Théry, T.; Steinberg, C.; Edel-Hermann, V.; Brand-Daunay, M.-C. Greenhouse Eggplant Production in France: Success and Failures of Grafting for Controlling Verticillium sp. In Proceedings of the 16. Eucarpia Capsicum and Eggplant Meeting, Kecskemét, Hungary, 12–14 September 2016; p. 6. [Google Scholar]
- Villeneuve, F.; Latour, F.; Théry, T.; Erard, P.; Fournier, C.; Daunay, M.C. Screening of Solanaceous Wild Relatives for Graft Affinity with Eggplant (Solanum melongena L.). In Proceedings of the 16. Eucarpia Capsicum and Eggplant Meeting, Kecskemét, Hungary, 12–14 September 2016; pp. 152–160. [Google Scholar]





| Geographic Region | Aubergine | Pepper | Tomato | Melon | Lettuce 1 |
|---|---|---|---|---|---|
| Australia and New Zealand | 59 | 4 | |||
| Melanesia | 2 | ||||
| Polynesia | 1 | ||||
| Caribbean | 75 | 79 | 26 | 18 | |
| Central America | 9 | 115 | 97 | 4 | |
| Northern America | 45 | 102 | 354 | 140 | 5 |
| South America | 29 | 170 | 208 | 21 | 2 |
| Central Asia | 4 | 3 | 36 | 5 | |
| Eastern Asia | 150 | 151 | 39 | 175 | 9 |
| Southeastern Asia | 237 | 52 | 4 | 19 | 1 |
| Southern Asia | 230 | 69 | 14 | 518 | |
| Western Asia | 51 | 57 | 378 | 370 | 18 |
| Eastern Africa | 195 | 50 | 6 | 7 | |
| Middle Africa | 34 | 22 | 12 | ||
| Northern Africa | 33 | 85 | 6 | 285 | 3 |
| Southern Africa | 60 | 4 | 3 | ||
| Western Africa | 406 | 41 | 20 | 8 | 1 |
| Eastern Europe | 53 | 222 | 175 | 109 | 1 |
| Northern Europe | 3 | 53 | 13 | 5 | |
| Southern Europe | 106 | 160 | 145 | 303 | 37 |
| Western Europe | 97 | 165 | 913 | 122 | 224 |
| Collection | Species/Crop Wild Relatives | Number of Species | Number of Accessions |
|---|---|---|---|
| Aubergine | Solanum melongena | 1211 | |
| S. aethiopicum | 335 | ||
| S. macrocarpon | 91 | ||
| Crop wild relatives—Solanum | 109 | 609 | |
| Other Solanaceae genus (8) | 17 | 24 | |
| Pepper | Capsicum annuum | 1683 | |
| C. baccatum | 129 | ||
| C. chinense | 159 | ||
| C. frutescens | 86 | ||
| C. pubescens | 28 | ||
| Crop wild relatives | 6 | 24 | |
| Tomato | Solanum lycopersicum | 3095 | |
| Crop wild relatives | 9 | 285 | |
| Melon | Cucumis melo 1 | 2359 | |
| Lettuce | Lactuca sativa | 712 | |
| Crop wild relatives—Lactuca | 10 | 225 | |
| Crop wild relatives—other genus | 3 | 15 |
| Plant Disease | Number of Accessions | Comment |
|---|---|---|
Bremia lactucae Reactive cultivars to stimulator of plant defences (SDP) for Bremia protection | 400 and 66 accessions within the European Evaluation Network project 402 cultivars tested with 3 SDP | EVA projet (2019–2023) https://www.ecpgr.cgiar.org/european-evaluation-network-eva/eva-networks/lettuce/ (accessed on 5 December 2021) Some reactive cultivars with good protection against Bremia [104] |
| Potyvirus lettuce mosaic virus Potyvirus lettuce Italian necrotic virus | 231 (116 cultivated and 115 wild) 20 (11 cultivated and 9 wild) | L. virosa PIVT1398 resistant to all lettuce mosaic virus strains [42]. One resistant: PIVT1398 [97] Same Mo3 locus, introgressed from L. virosa, confers resistance to LMV and to LINV [43] |
| Xanthomonas campestris | 986 (789 cultivated and 197 wild) | Few genitors in cultivars [99] QTL analysis in RIL population [105] |
| Meloidogyne incognita | 569 (409 cultivated and 160 wild) | Resistance found in L. sativa and L. serriola [100] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Salinier, J.; Lefebvre, V.; Besombes, D.; Burck, H.; Causse, M.; Daunay, M.-C.; Dogimont, C.; Goussopoulos, J.; Gros, C.; Maisonneuve, B.; et al. The INRAE Centre for Vegetable Germplasm: Geographically and Phenotypically Diverse Collections and Their Use in Genetics and Plant Breeding. Plants 2022, 11, 347. https://doi.org/10.3390/plants11030347
Salinier J, Lefebvre V, Besombes D, Burck H, Causse M, Daunay M-C, Dogimont C, Goussopoulos J, Gros C, Maisonneuve B, et al. The INRAE Centre for Vegetable Germplasm: Geographically and Phenotypically Diverse Collections and Their Use in Genetics and Plant Breeding. Plants. 2022; 11(3):347. https://doi.org/10.3390/plants11030347
Chicago/Turabian StyleSalinier, Jérémy, Véronique Lefebvre, Didier Besombes, Hélène Burck, Mathilde Causse, Marie-Christine Daunay, Catherine Dogimont, Juliette Goussopoulos, Christophe Gros, Brigitte Maisonneuve, and et al. 2022. "The INRAE Centre for Vegetable Germplasm: Geographically and Phenotypically Diverse Collections and Their Use in Genetics and Plant Breeding" Plants 11, no. 3: 347. https://doi.org/10.3390/plants11030347
APA StyleSalinier, J., Lefebvre, V., Besombes, D., Burck, H., Causse, M., Daunay, M. -C., Dogimont, C., Goussopoulos, J., Gros, C., Maisonneuve, B., McLeod, L., Tobal, F., & Stevens, R. (2022). The INRAE Centre for Vegetable Germplasm: Geographically and Phenotypically Diverse Collections and Their Use in Genetics and Plant Breeding. Plants, 11(3), 347. https://doi.org/10.3390/plants11030347