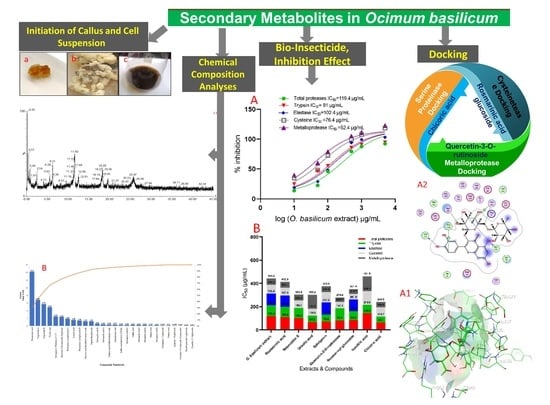
Secondary Metabolites in Basil, Bio-Insecticide, Inhibition Effect, and In Silico Molecular Docking against Proteolytic Enzymes of the Red Palm Weevil (Rhynchophorus ferrugineus)
Abstract
:1. Introduction
2. Results
2.1. Initiation of Callus and Cell Suspension
2.2. Chemical Content and Chemical Composition Analyses of O. basilicum Extract
2.3. Polyphenolic Acids and Flavonoids Compounds in O. basilicum Cell Suspension Extracts Using UPLC–I Class Coupled with Xevo TQD MS
2.4. O. basilicum Extract and Pure Compounds Activity against Adults and Larvae of R. ferrugineus
2.5. Evaluation Specific Activity of O. basilicum Extract and Pure Compounds on Serine, Cysteine, and Metalloproteinase (In Vitro)
2.6. In Vivo Effect of Specific Protease Inhibitors and O. basilicum Extract on the Serine, Metalloprotease, and Cysteine Protease Activities from Fourth R. ferrugineus Instar Midgut Preparations
2.7. Docking of Compounds into Proteinase Enzymes
2.7.1. Serine Proteinase Docking
2.7.2. Cysteine Protease Docking
2.7.3. Metalloprotease Docking
2.8. ADMET Analysis
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Media
4.3. Plants
4.4. O. basilicum Calluses Initiation Employing Various PGRs in Conjunction and V. dahliae as a Biotic Elicitor
4.5. O. basilicum Cell Suspension Initiation
4.6. Total Phenolic Content (TPC) Determination
4.7. Total Flavonoids Contents (TF)
4.8. Determination of Total Condensed Tannins (TCT)
4.9. Liquid Chromatography-Mass Spectrometry Analysis (LC-MS)
4.10. Evaluation of the Extracted Secondary Metabolites’ Contact-Insecticide and Antifeedant Efficacy against R. ferrugineus
4.11. Assessment of an O. basilicum Cell Suspension Extract and Pure Components on the Proteolytic Enzyme Activity (In Vitro) of R. ferrugineus Larvae
4.12. Assessment of an O. basilicum Cell Suspension Extract and Pure Components on Serine Proteinase Specific Activity (In Vitro) of R. ferrugineus Larvae
4.13. Assessment of an O. basilicum Cell Suspension Extract and Pure Components on Metalloproteinase Specific Activity (In Vitro) of R. ferrugineus Larvae
4.14. Assessment of an O. basilicum Cell Suspension Extract and Pure Components on Cysteine Proteinase Specific Activity (In Vitro) of R. ferrugineus Larvae
4.15. Docking of Tested Compounds into Enzymes
4.16. ADMET Screening
4.17. Statistical Design
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darrag, H.M.; Alhajhoj, M.R.; Khalil, H.E. Bio-Insecticide of Thymus vulgaris and Ocimum basilicum Extract from Cell Suspensions and Their Inhibitory Effect against Serine, Cysteine, and Metalloproteinases of the Red Palm Weevil (Rhynchophorus ferrugineus). Insects 2021, 12, 405. [Google Scholar] [CrossRef] [PubMed]
- Milosavljević, I.; El-Shafie, H.A.; Faleiro, J.R.; Hoddle, C.D.; Lewis, M.; Hoddle, M.S. Palmageddon: The wasting of ornamental palms by invasive palm weevils, Rhynchophorus spp. J. Pest Sci. 2019, 92, 143–156. [Google Scholar] [CrossRef]
- Shehawy, A.A.; Ibrahim, M.T.; Aboutaleb, E.S.; Qari, S.H. Bioactivity and biochemical efficacy of chitinase and Justicia brandegeana extract against Red Palm Weevil Rhynchophorus ferrugineus Olivier (Coleoptera: Curculionidae). Food Sci. Nutr. 2020, 8, 4625–4636. [Google Scholar] [CrossRef] [PubMed]
- Idris, A.M.; Miller, T.A.; Durvasula, R.; Fedoroff, N. Bridging the Knowledge Gaps for Development of Basic Components of Red Palm Weevil IPM. In Sustainable Pest Management in Date Palm: Current Status and Emerging Challenges; Springer: New York, NY, USA, 2015; pp. 37–62. [Google Scholar]
- Faleiro, J. A review of the issues and management of the red palm weevil Rhynchophorus ferrugineus (Coleoptera: Rhynchophoridae) in coconut and date palm during the last one hundred years. Int. J. Trop. Insect Sci. 2006, 26, 135–154. [Google Scholar]
- Downer, A.J.; Uchida, J.Y.; Hodel, D.R.; Elliott, M.L. Lethal palm diseases common in the United States. HortTechnology 2009, 19, 710–716. [Google Scholar] [CrossRef] [Green Version]
- Kontodimas, D.; Soroker, V.; Pontikakos, C.; Suma, P.; Beaudoin-Ollivier, L.; Karamaouna, F.; Riolo, P. Visual Identification and Characterization of Rhynchophorus ferrugineus and Paysandisia archon Infestation. In Handbook of Major Palm Pests: Biology and Management; Wiley: Hoboken, NJ, USA, 2016; pp. 187–208. [Google Scholar]
- Wattanapongsiri, A. A Revision of the Genera Rhynchophorus and Dynamis (Coleoptera: Curculionidae). Ph.D. Dissertation, Oregon State University Corvallis, Corvallis, OR, USA, 1966. [Google Scholar]
- Žďárek, J.; Howard, F.W.; Moore, D.; Giblin-Davis, R.M.; Abad, R.G. Insects on Palms.(Ecological Studies 142.). Biol. Plant. 2002, 45, 196. [Google Scholar] [CrossRef]
- Fiaboe, K.; Peterson, A.T.; Kairo, M.; Roda, A. Predicting the potential worldwide distribution of the red palm weevil Rhynchophorus ferrugineus (Olivier) (Coleoptera: Curculionidae) using ecological niche modeling. Fla. Entomol. 2012, 95, 659–673. [Google Scholar] [CrossRef]
- Rugman-Jones, P.F.; Hoddle, C.D.; Hoddle, M.S.; Stouthamer, R. The lesser of two weevils: Molecular-genetics of pest palm weevil populations confirm Rhynchophorus vulneratus (Panzer 1798) as a valid species distinct from R. ferrugineus (Olivier 1790), and reveal the global extent of both. PLoS ONE 2013, 8, e78379. [Google Scholar] [CrossRef]
- Hoddle, M.; Hoddle, C. Palmageddon: The invasion of California by the South American palm weevil is underway. CAPCA Advis. 2017, 20, 40–44. [Google Scholar]
- Ahmed, F.; Hussein, K.; Gad, M. Biological activity of four plant oils, against the red palm weevil, Rhynchophorus ferrugineus (Oliver), (Coleoptera: Curculionidae). J. Biosci. Appl. Res. 2015, 1, 213–222. [Google Scholar] [CrossRef]
- Abdel-Raheem, M.; ALghamdi, H.A.; Reyad, N.F. Nano essential oils against the red palm weevil, Rhynchophorus ferrugineus Olivier (Coleoptera: Curculionidae). Entomol. Res. 2020, 50, 215–220. [Google Scholar] [CrossRef]
- Cangelosi, B.; Clematis, F.; Monroy, F.; Roversi, P.F.; Troiano, R.; Curir, P.; Lanzotti, V. Filiferol, a chalconoid analogue from Washingtonia filifera possibly involved in the defence against the Red Palm Weevil Rhynchophorus ferrugineus Olivier. Phytochemistry 2015, 115, 216–221. [Google Scholar] [CrossRef] [PubMed]
- Orfali, R.; Binsuwaileh, A.; Al-Ala’a, H.A.; Bane-Gamea, S.; Zaidan, N.; Abdelazim, M.; Ismael, M.A.; Perveen, S.; Majrashi, N.; Alluhayb, K. Production of a biopesticide on host and Non-Host serine protease inhibitors for red palm weevil in palm trees. Saudi J. Biol. Sci. 2020, 27, 2803–2808. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Sifuentes, L.; Marszalekfer, J.E.; Chuck-Hernández, C.; Serna-Saldívar, S.O. Legumes Protease Inhibitors as Biopesticides and Their Defense Mechanisms against Biotic Factors. Int. J. Mol. Sci. 2020, 21, 3322. [Google Scholar] [CrossRef]
- Saad, M.M.; Gouda, N.A.; Abdelgaleil, S.A. Bioherbicidal activity of terpenes and phenylpropenes against Echinochloa crus-galli. J. Environ. Sci. Health Part B 2019, 54, 954–963. [Google Scholar] [CrossRef]
- Guarino, S.; Colazza, S.; Peri, E.; Bue, P.L.; Germanà, M.P.; Kuznetsova, T.; Gindin, G.; Soroker, V. Behaviour-modifying compounds for management of the red palm weevil (Rhynchophorus ferrugineus Oliver). Pest Manag. Sci. 2015, 71, 1605–1610. [Google Scholar] [CrossRef]
- Guarino, S.; Peri, E.; Bue, P.L.; Germanà, M.P.; Colazza, S.; Anshelevich, L.; Ravid, U.; Soroker, V. Assessment of synthetic chemicals for disruption of Rhynchophorus ferrugineus response to attractant-baited traps in an urban environment. Phytoparasitica 2013, 41, 79–88. [Google Scholar] [CrossRef]
- AlJabr, A.M.; Hussain, A.; Rizwan-ul-Haq, M.; Al-Ayedh, H. Toxicity of plant secondary metabolites modulating detoxification genes expression for natural red palm weevil pesticide development. Molecules 2017, 22, 169. [Google Scholar] [CrossRef] [Green Version]
- Hussain, A.; Rizwan-Ul-Haq, M.; AlJabr, A.M.; Al-Ayedh, H. Lethality of sesquiterpenes reprogramming red palm weevil detoxification mechanism for natural novel biopesticide development. Molecules 2019, 24, 1648. [Google Scholar] [CrossRef] [Green Version]
- Prinsi, B.; Morgutti, S.; Negrini, N.; Faoro, F.; Espen, L. Insight into Composition of Bioactive Phenolic Compounds in Leaves and Flowers of Green and Purple Basil. Plants 2020, 9, 22. [Google Scholar] [CrossRef] [Green Version]
- Elansary, H.O.; Szopa, A.; Kubica, P.; Ekiert, H.; El-Ansary, D.O.; Al-Mana, F.A.; Mahmoud, E.A. Saudi Rosmarinus officinalis and Ocimum basilicum L. Polyphenols and Biological Activities. Processes 2020, 8, 446. [Google Scholar] [CrossRef] [Green Version]
- Wojdyło, A.; Oszmiański, J.; Czemerys, R. Antioxidant activity and phenolic compounds in 32 selected herbs. Food Chem. 2007, 105, 940–949. [Google Scholar] [CrossRef]
- Al-Asmari, A.K.; Athar, M.T.; Al-Faraidy, A.A.; Almuhaiza, M.S. Chemical composition of essential oil of Thymus vulgaris collected from Saudi Arabian market. Asian Pac. J. Trop. Biomed. 2017, 7, 147–150. [Google Scholar] [CrossRef]
- Chenni, M.; El Abed, D.; Rakotomanomana, N.; Fernandez, X.; Chemat, F. Comparative study of essential oils extracted from Egyptian basil leaves (Ocimum basilicum L.) using hydrodistillation and solvent-free microwave extraction. Molecules 2016, 21, 113. [Google Scholar] [CrossRef] [Green Version]
- Padalia, R.; Verma, R.; Chauhan, A.; Chanotiya, C. Changes in aroma profiles of 11 Indian Ocimum taxa during plant ontogeny. Acta Physiol. Plant. 2013, 35, 2567–2587. [Google Scholar] [CrossRef]
- Murthy, H.N.; Lee, E.-J.; Paek, K.-Y. Production of secondary metabolites from cell and organ cultures: Strategies and approaches for biomass improvement and metabolite accumulation. Plant Cell Tissue Organ Cult. (PCTOC) 2014, 118, 1–16. [Google Scholar] [CrossRef]
- Monfort, L.E.F.; Bertolucci, S.K.V.; Lima, A.F.; de Carvalho, A.A.; Mohammed, A.; Blank, A.F.; Pinto, J.E.B.P. Effects of plant growth regulators, different culture media and strength MS on production of volatile fraction composition in shoot cultures of Ocimum basilicum. Ind. Crops Prod. 2018, 116, 231–239. [Google Scholar] [CrossRef]
- Ramachandra Rao, S.; Ravishankar, G.A. Plant cell cultures: Chemical factories of secondary metabolites. Biotechnol. Adv. 2002, 20, 101–153. [Google Scholar] [CrossRef]
- Açıkgöz, M.A. Establishment of cell suspension cultures of Ocimum basilicum L. and enhanced production of pharmaceutical active ingredients. Ind. Crops Prod. 2020, 148, 112278. [Google Scholar] [CrossRef]
- Khalil, H.E.; Ibrahim, H.I.; Darrag, H.M.; Matsunami, K. Insight into Analysis of Essential Oil from Anisosciadium lanatum Boiss.—Chemical Composition, Molecular Docking, and Mitigation of Hepg2 Cancer Cells through Apoptotic Markers. Plants 2021, 11, 66. [Google Scholar] [CrossRef]
- Khalil, H.E.; Alqahtani, N.K.; Darrag, H.M.; Ibrahim, H.-I.M.; Emeka, P.M.; Badger-Emeka, L.I.; Matsunami, K.; Shehata, T.M.; Elsewedy, H.S. Date Palm Extract (Phoenix dactylifera) PEGylated Nanoemulsion: Development, Optimization and Cytotoxicity Evaluation. Plants 2021, 10, 735. [Google Scholar] [CrossRef]
- Ibrahim, H.-I.M.; Darrag, H.M.; Alhajhoj, M.R.; Khalil, H.E. Biomolecule from Trigonella stellata from Saudi Flora to Suppress Osteoporosis via Osteostromal Regulations. Plants 2020, 9, 1610. [Google Scholar] [CrossRef] [PubMed]
- Younis, H.M.; Badawy, M.E.; Darrag, H.M. Tissue culture of the Egyptian cotton cultivars: Production and morphological heterogeneity of primary callus tissues. Exp. Biol. 2013, 27, 1014.1. [Google Scholar] [CrossRef]
- Lee, J.; Chan, B.L.S.; Mitchell, A.E. Identification/Quantification of Free and Bound Phenolic Acids in Peel and Pulp of Apples (Malus domestica) Using High Resolution Mass Spectrometry (HRMS). Food Chem. 2017, 215, 301–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, L.; Fu, S.; Du, W.; Wang, B.; Li, L.; Zhu, M.; Liu, C.; Zhang, J. Validation and Application of an Rapid HPLC–MS Method for the Determination of Salvianic Acid A in Human Plasma. J. Chromatogr. Sci. 2015, 53, 771–777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoza, B.S.; Gbashi, S.; Steenkamp, P.A.; Njobeh, P.B.; Madala, N.E. Identification of Hydroxylcinnamoyl Tartaric Acid Esters in Bidens Pilosa by UPLC-Tandem Mass Spectrometry. S. Afr. J. Bot. 2016, 103, 95–100. [Google Scholar] [CrossRef]
- Ruan, M.; Li, Y.; Li, X.; Luo, J.; Kong, L. Qualitative and Quantitative Analysis of the Major Constituents in Chinese Medicinal Preparation Guan-Xin-Ning Injection by HPLC–DAD–ESI-MSn. J. Pharm. Biomed. Anal. 2012, 59, 184–189. [Google Scholar] [CrossRef]
- Lu, Y.; Foo, Y. Rosmarinic Acid Derivatives from Salvia officinalis. Phytochemistry 1999, 51, 91–94. [Google Scholar] [CrossRef]
- Barros, L.; Dueñas, M.; Dias, M.I.; Sousa, M.J.; Santos-Buelga, C.; Ferreira, I.C.F.R. Phenolic Profiles of Cultivated, In Vitro Cultured and Commercial Samples of Melissa officinalis L. Infusions. Food Chem. 2013, 136, 1–8. [Google Scholar] [CrossRef]
- Prinsi, B.; Negri, A.S.; Quattrocchio, F.M.; Koes, R.E.; Espen, L. Proteomics of Red and White Corolla Limbs in Petunia Reveals a Novel Function of the Anthocyanin Regulator ANTHOCYANIN1 in Determining Flower Longevity. J. Proteom. 2016, 131, 38–47. [Google Scholar] [CrossRef]
- Abad-García, B.; Garmón-Lobato, S.; Berrueta, L.A.; Gallo, B.; Vicente, F. A Fragmentation Study of Dihydroquercetin Using Triple Quadrupole Mass Spectrometry and Its Application for Identification of Dihydroflavonols in Citrus Juices. Rapid Commun. Mass Spectrom. 2009, 23, 2785–2792. [Google Scholar] [CrossRef] [PubMed]
- Nováková, L.; Vildová, A.; Mateus, J.P.; Gonçalves, T.; Solich, P. Development and Application of UHPLC–MS/MS Method for the Determination of Phenolic Compounds in Chamomile Flowers and Chamomile Tea Extracts. Talanta 2010, 82, 1271–1280. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.B.; Rai, D.K.; Brunton, N.P.; Martin-Diana, A.B.; Barry-Ryan, C. Characterization of phenolic composition in Lamiaceae spices by LC-ESI-MS/MS. J. Agric. Food Chem. 2010, 58, 10576–10581. [Google Scholar] [CrossRef] [PubMed]
- Mena, P.; Calani, L.; Dall’Asta, C.; Galaverna, G.; García-Viguera, C.; Bruni, R.; Crozier, A.; Del Rio, D. Rapid and Comprehensive Evaluation of (Poly)phenolic Compounds in Pomegranate (Punica granatum L.) Juice by UHPLC-MSn. Molecules 2012, 17, 14821–14840. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Li, X.; Saleri, F.; Guo, M. Analysis of Flavonoids in Rhamnus davurica and Its Antiproliferative Activities. Molecules 2016, 21, 1275. [Google Scholar] [CrossRef]
- Stander, M.A.; Van Wyk, B.E.; Taylor, M.; Long, H.S. Analysis of Phenolic Compounds in Rooibos Tea (Aspalathus linearis) with a Comparison of Flavonoid-Based Compounds in Natural Populations of Plants from Different Regions. J. Agric. Food Chem. 2017, 65, 10270. [Google Scholar] [CrossRef]
- Ben Said, R.; Hamed, A.I.; Mahalel, U.A.; Al-Ayed, A.S.; Kowalczyk, M.; Moldoch, J.; Oleszek, W.; Stochmal, A. Tentative Characterization of Polyphenolic Compounds in the Male Flowers of Phoenix dactylifera by Liquid Chromatography Coupled with Mass Spectrometry and DFT. Int. J. Mol. Sci. 2017, 18, 512. [Google Scholar] [CrossRef]
- Ye, M.; Guo, D.; Ye, G.; Huang, C. Analysis of homoisoflavonoids in Ophiopogon japonicus by HPLC-DAD-ESI-MS. J. Am. Soc. Mass Spectrom. 2005, 16, 234–243. [Google Scholar] [CrossRef] [Green Version]
- Ross, D.C.; Brown, T.M. Inhibition of larval growth in Spodoptera frugiperda by sublethal dietary concentrations of insecticides. J. Agric. Food Chem. 1982, 30, 193–196. [Google Scholar] [CrossRef]
- Lewis, N.G.; Yamamoto, E. Lignin: Occurrence, biogenesis and biodegradation. Annu. Rev. Plant Biol. 1990, 41, 455–496. [Google Scholar] [CrossRef]
- Bolwell, G.P.; Robbins, M.P.; Dixon, R.A. Metabolic changes in elicitor-treated bean cells: Enzymic responses associated with rapid changes in cell wall components. Eur. J. Biochem. 1985, 148, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Kefeli, V.I.; Kalevitch, M.V.; Borsari, B. Phenolic cycle in plants and environment. J. Cell Mol. Biol. 2003, 2, 13–18. [Google Scholar]
- López Arnaldos, T.; Muñoz, R.; Ferrer, M.A.; Calderón, A.A. Changes in phenol content during strawberry (Fragaria × ananassa, cv. Chandl. ) callus culture. Physiol. Plant. 2001, 113, 315–322. [Google Scholar]
- Mato, M.; Rua, M.; Ferro, a. Changes in levels of peroxidases and phenolics during root formation in Vitis cultured in vitro. Physiol. Plant. 1988, 72, 84–88. [Google Scholar] [CrossRef]
- Pickens, C.L.; Airavaara, M.; Theberge, F.; Fanous, S.; Hope, B.T.; Shaham, Y. Neurobiology of the incubation of drug craving. Trends Neurosci. 2011, 34, 411–420. [Google Scholar] [CrossRef] [Green Version]
- Atanasov, A.G.; Waltenberger, B.; Pferschy-Wenzig, E.-M.; Linder, T.; Wawrosch, C.; Uhrin, P.; Temml, V.; Wang, L.; Schwaiger, S.; Heiss, E.H. Discovery and resupply of pharmacologically active plant-derived natural products: A review. Biotechnol. Adv. 2015, 33, 1582–1614. [Google Scholar] [CrossRef] [Green Version]
- Ochoa-Villarreal, M.; Howat, S.; Hong, S.; Jang, M.O.; Jin, Y.-W.; Lee, E.-K.; Loake, G.J. Plant cell culture strategies for the production of natural products. BMB Rep. 2016, 49, 149. [Google Scholar] [CrossRef]
- Mathew, R.; Sankar, P.D. Comparison of major secondary metabolites quantified in elicited cell cultures, non-elicited cell cultures, callus cultures and field grown plants of Ocimum. Int. J. Pharm. Pharm. Sci. 2014, 6, 102–106. [Google Scholar]
- Michaud, N.R.; Fabian, J.R.; Mathes, K.D.; Morrison, D.K. 14-3-3 is not essential for Raf-1 function: Identification of Raf-1 proteins that are biologically activated in a 14-3-3-and Ras-independent manner. Mol. Cell. Biol. 1995, 15, 3390–3397. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Child, M.A.; Bogyo, M. Proteases as regulators of pathogenesis: Examples from the Apicomplexa. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2012, 1824, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Gomes, M.T.; Teixeira, R.D.; Lopes, M.T.; Nagem, R.A.; Salas, C.E. X-ray crystal structure of CMS1MS2: A high proteolytic activity cysteine proteinase from Carica candamarcensis. Amino Acids 2012, 43, 2381–2391. [Google Scholar] [CrossRef] [PubMed]
- Waterman, P.G.; Mole, S. Analysis of Phenolic Plant Metabolites; Blackwell Scientific Oxford: Oxfordshire, UK, 1994; Volume 83. [Google Scholar]
- Waterhouse, A.L. Determination of Total Phenolics. Curr. Protoc. Food Anal. Chem. 2002, 6, I1.1.1. [Google Scholar]
- Hagerman, A. Quantification of Tannins in Tree Foliage: A Laboratory Manual for the FAO/IAEA Co-Ordinated Research Project on the Use of Nuclear and Related Techniques to Develop Simple Tannin Assays for Predicting and Improving the Safety and Efficiency of Feeding Ruminants on Tanninferous Tree Foliage; IAEA Division of Nuclear Techniques in Food and Agriculture: Vienna, Austria, 2000; Volume 33, pp. 1–26. Available online: http://www.iaea.org/programmes/nafa/dx/ (accessed on 27 March 2022).
- Shukla, P.; Vidyasagar, P.; Aldosari, S.A.; Abdel-Azim, M. Antifeedant activity of three essential oils against the red palm weevil, Rhynchophorus ferrugineus. Bull. Insectology 2012, 65, 71–76. [Google Scholar]
- Lowry, O.; Rosebrough, N.J.; Farr, A.; Randall, R.J. Protein Measurement with the Folin Phenol Reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Olga, L.; Ibrahim, M.; Candas, N.; Koller, N.; Bauer, L.; Bulla, L. Changes in proteases activity and cry 3Aa toxin binding in the Colorado potato beetle: Implications for insect resistance to Bacillus thuringiensis toxins. Insect Biochem. Mol. Biol. 2002, 32, 567–577. [Google Scholar]
- Kawatkar, S.P.; Kuntz, D.A.; Woods, R.J.; Rose, D.R.; Boons, G.J. Structural basis of the inhibition of Golgi α-mannosidase II by mannostatin A and the role of the thiomethyl moiety in ligand−protein interactions. J. Am. Chem. Soc. 2006, 128, 8310–8319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumann, U.; Wu, S.; Flaherty, K.M.; McKay, D.B. Three-dimensional structure of the alkaline protease of Pseudomonas aeruginosa: A two-domain protein with a calcium binding parallel beta roll motif. EMBO J. 1993, 12, 3357–3364. [Google Scholar] [CrossRef]
- Lipinski, C.A. Lead-and drug-like compounds: The rule of-five revolution. Drug Discov. Today Technol. 2004, 1, 337–341. [Google Scholar] [CrossRef]
- Finney, D.J. Probit Analysis; Cambridge University Press: Cambridge, UK, 1971. [Google Scholar]







| TF (mg of Quercetin/g DW) | TPC (mg of Gallic Acid/g DW) | Callus and Cell Suspension | TCT (mg of Cyanidins/g DW) |
|---|---|---|---|
| 0.95 d ± 0.1324 | 9.78 d ± 0.1109 | Callus without infection | 0.24 d ± 0.0454 |
| 1.36 c ± 0.1562 | 14.85 c ± 0.1674 | Callus with infection | 0.38 c ± 0.0742 |
| 2.21 b ± 0.0413 | 19.23 b ± 0.1457 | Cell suspension without infection | 0.41 b ± 0.0316 |
| 3.97 a ± 0.0478 | 32.51 a ± 0.1904 | Cell suspension with infection | 0.74 a ± 0.1245 |
| No. | Compounds Tentatively | RT (min) | RI (exp) | Formula | [M − H]− (m/z) | Fragmentation Ions (m/z) | Content (µmol g−1 cell) |
|---|---|---|---|---|---|---|---|
| 1 | Tartaric acid | 1.01 | 1249 | C4H5O6 | 149.00 | 149, 141, 131, 113, 103, 87 | 0.18 |
| 2 | Isocitric acid | 2.54 | 1805.4 | C6H7O7 | 191.0175 | 191, 173, 129, 111 | 1.58 |
| 3 | Caffeic acid derivative (3TMS) | 4.13 | 2155 | C18H32O4Si3 | 359.70 | 396, 381, 359, 219, 191, 75 | 0.28 |
| 4 | Caftaric acid (Caffeoyl-tartaric acid) | 5.63 | 2701.3 | C13H12O9 | 311.04 | 311, 179.03, 149.01, 135.04 | 0.38 |
| 5 | Caffeic acid | 6.28 | 1854.3 | C9H8O4 | 179.03 | 179, 135 | 0.23 |
| 6 | Fertaric acid | 6.31 | 5191.1 | C14H14O9 | 325.06 | 325, 193, 134 | 0.12 |
| 7 | Salvianolic acid H/I | 6.38 | 5237.8 | C27H22O12 | 537.10 | 537, 493, 339, 313, 295, 197, 179 | 0.28 |
| 8 | Salvianolic acid K | 9.57 | 4556.9 | C27H24O13 | 555.11 | 555, 537, 493, 295 | 0.22 |
| 9 | Chicoric acid (dicaffeoyl-tartaric acid) | 11.18 | 4552.3 | C22H18O12 | 473.07 | 473, 311, 293, 179, 149 | 1.23 |
| 10 | Lithospermic acid | 11.31 | 4920.2 | C27H22O12 | 537.10 | 537, 493, 356, 295 | 0.37 |
| 11 | Dihydroquercetin 3-glucoside | 11.46 | 4505.7 | C21H22O12 | 456.10 | 467, 465, 313, 285, 259, 456, 175, 151 | 0.021 |
| 12 | Quercetin-3-O-rutinoside (rutin) | 11.53 | 4992.3 | C27H30O16 | 611.16 | 611, 465, 449, 303 | 2.34 |
| 12 | Rosmarinic acid | 11.60 | 3504.5 | C18H16O8 | 359.08 | 359, 197, 179, 161, 135, 117 | 14.2 |
| 13 | Salvianolic acid E | 12.69 | 4627.5 | C36H30O16 | 717.15 | 717, 519, 475, 339 | 0.14 |
| 14 | Salvianolic acid A | 12.49 | 4585.8 | C26H22O10 | 493.11 | 493, 313, 295, 185 | 0.42 |
| 15 | Salvianolic acid B | 12.61 | 5377.7 | C36H30O16 | 717.15 | 717, 519, 321 | 0.54 |
| 16 | Salvianolic acid F | 17.94 | 4566.3 | C17H14O6 | 313.07 | 313, 269 | 0.23 |
| 17 | Cyanidin 3,3’-diglucoside | 18.14 | 6158.2 | C27H31O16 | 611.16 | 611, 287 | 0.021 |
| 18 | Cyanidin 3-O-rutinoside (Cyaninoside) | 18.27 | 5192.3 | C27H31O15 | 595.17 | 595, 287 | 0.027 |
| 19 | Salvigenin (5-Hydroxy-6,7,4′-trimethoxyflavone) | 18.29 | 3121.7 | C18H16O6 | 327.21 | 327, 311, 277, 215, 205, 116.9 | 2.51 |
| 20 | Naringenin 7-0-glucoside | 18.36 | 4081.3 | C21H22Os10 | 434.4 | 435, 271, 151, 119 | 0.123 |
| 21 | Apigenin 7-O-glucoside | 18.45 | 4142.7 | C21H20O10 | 432.4 | 432, 271, 171, 147, 119 | 0.078 |
| 22 | Rosmarinic acid glucoside A | 21.37 | 4023.4 | C24H26O13 | 521.12 | 359, 197, 179, 161, 135 | 1.87 |
| 23 | Rosmarinic acid glucoside B | 25.07 | 4061.4 | C24H26O13 | 521.12 | 359, 323, 197, 179, 161, 135 | 1.45 |
| 24 | Nepetoidin A | 25.53 | 4413.7 | C17H14O6 | 314.29 | 335, 313, 161, 133 | 6.84 |
| 25 | Nepetoidin B | 25.67 | 4418.9 | C17H14O6 | 314.29 | 335, 313, 269, 161, 133 | 5.72 |
| 26 | Ursolic acid | 26.06 | 3658.3 | C30H48O3 | 456.7 | 591, 524, 523, 459, 455 | 4.91 |
| 27 | Nepetoidin glucoside | 27.99 | 4341 | C23H24O11 | 475.12 | 475, 323, 313, 161, 151 | 1.23 |
| 28 | Unknown | 36.11 | 3697.2 | ND | ND | ND | ND |
| 29 | Unknown | 36.80 | 3751 | ND | ND | ND | ND |
| 30 | Unknown | 42.34 | 3508 | ND | ND | ND | ND |
| Extract and Compounds | Adult | 4th Larvae | ||||||
|---|---|---|---|---|---|---|---|---|
| LC50 (µg/mL) | Slope | Chi Square | p | LD50 (µg/Larvae) | Slope | Chi Square | p | |
| O. basilicum extract | 1238 (1038–1389) | 2.64 ± 0.20 | 48.41 | 0.003 | 13.7 (12.9–15.6) | 1.84 ± 0.26 | 43.42 | 0.001 |
| Rosmarinic acid | 1495 (1378–1504) | 2.61 ± 0.22 | 42.63 | 0.007 | 12.4 (11.8–12.7) | 1.94 ± 0.25 | 45.23 | 0.005 |
| Nepetoidin B | 1317 (1268–1346) | 2.82 ± 0.20 | 43.29 | 0.004 | 11.9 (11.1–12.3) | 1.97 ± 0.23 | 46.78 | 0.003 |
| Ursolic acid | 1167 (1038–1204) | 2.85 ± 0.23 | 43.85 | 0.004 | 15.2 (14.2–15.9) | 1.46 ± 0.28 | 41.21 | 0.003 |
| Salvigenin | 1189 (1049–1219) | 2.87 ± 0.20 | 44.27 | 0.005 | 11.4 (10.4–11.8) | 2.03 ± 0.22 | 47.54 | 0.004 |
| Quercetin-3-O-rutinoside | 1214 (1089–1234) | 2.89 ± 0.24 | 42.36 | 0.003 | 16.9 (15.1–17.6) | 1.42 ± 0.29 | 42.31 | 0.002 |
| Rosmarinyl glucoside | 1275 (1147–1315) | 2.91 ± 0.21 | 40.85 | 0.003 | 17.6 (15.8–18.3) | 1.40 ± 0.31 | 43.98 | 0.002 |
| Isocitric acid | 1826 (1712–1976) | 2.12 ± 0.19 | 49.29 | 0.009 | 23.9 (21.1–24.6) | 1.08 ± 0.30 | 42.36 | 0.008 |
| Chicoric acid | 1132 (1004–1198) | 2.97 ± 0.21 | 44.17 | 0.003 | 10.23 (9.87–10.94) | 2.07 ± 0.22 | 48.72 | 0.002 |
| Compounds | Docking Score ΔG (kcal/mol) | ||
|---|---|---|---|
| Serine Proteinase | Cysteine Protease | Metalloprotease | |
| Rosmarinic acid | −6.3212 | −6.4056 | −5.7938 |
| Nepetoidin A | −5.9404 | −5.3541 | −6.0523 |
| Nepetoidin B | −6.0265 | −5.4078 | −5.8765 |
| Ursolic acid | −4.9541 | −6.4766 | −6.6226 |
| Salvigenin | −6.2783 | −5.3786 | −6.2654 |
| Quercetin-3-O-rutinoside | −8.1833 | −6.5665 | −5.6609 |
| Rosmarinic acid glucoside | −7.7259 | −6.5869 | −6.0175 |
| Isocitric acid | −4.3629 | −4.1368 | −4.4249 |
| Chicoric acid | −6.3303 | −5.8215 | −6.8202 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Darrag, H.M.; Almuhanna, H.T.; Hakami, E.H. Secondary Metabolites in Basil, Bio-Insecticide, Inhibition Effect, and In Silico Molecular Docking against Proteolytic Enzymes of the Red Palm Weevil (Rhynchophorus ferrugineus). Plants 2022, 11, 1087. https://doi.org/10.3390/plants11081087
Darrag HM, Almuhanna HT, Hakami EH. Secondary Metabolites in Basil, Bio-Insecticide, Inhibition Effect, and In Silico Molecular Docking against Proteolytic Enzymes of the Red Palm Weevil (Rhynchophorus ferrugineus). Plants. 2022; 11(8):1087. https://doi.org/10.3390/plants11081087
Chicago/Turabian StyleDarrag, Hossam Moustafa, Hani Taher Almuhanna, and Emadaldeen Hamad Hakami. 2022. "Secondary Metabolites in Basil, Bio-Insecticide, Inhibition Effect, and In Silico Molecular Docking against Proteolytic Enzymes of the Red Palm Weevil (Rhynchophorus ferrugineus)" Plants 11, no. 8: 1087. https://doi.org/10.3390/plants11081087
APA StyleDarrag, H. M., Almuhanna, H. T., & Hakami, E. H. (2022). Secondary Metabolites in Basil, Bio-Insecticide, Inhibition Effect, and In Silico Molecular Docking against Proteolytic Enzymes of the Red Palm Weevil (Rhynchophorus ferrugineus). Plants, 11(8), 1087. https://doi.org/10.3390/plants11081087