The Invasive Tradescantia zebrina Affects Litter Decomposition, but It Does Not Change the Lignocellulolytic Fungal Community in the Atlantic Forest, Brazil
Abstract
:1. Introduction
2. Results
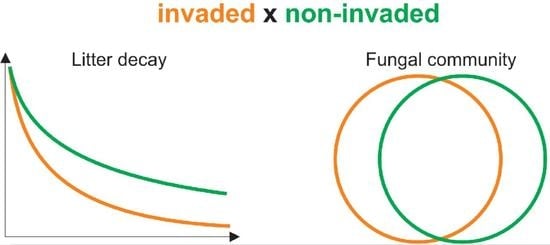
2.1. Decomposition of Litter under Different Environmental Conditions
2.2. Community of Decomposing Fungi
3. Discussion
4. Materials and Methods
4.1. Description of the Study Area
4.2. T. zebrine
4.3. Litter Bag Preparation
4.4. In Situ Experiment
4.5. In Vitro Experiment
4.6. Litterbags Processing
4.7. Isolation and Identification of Decomposing Fungi from Plant Litter
4.8. Analysis of the Community of Fungal Decomposers
4.9. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berg, B.; McClaugherty, C. Plant Litter: Decomposition, Humus Formation, Carbon Sequestration, 3rd ed.; Springer: Berlin/Heidelberg, Germany, 2014; p. 317. [Google Scholar]
- Krishna, M.P.; Mohan, M. Litter decomposition in forest ecosystems: A review. Energy Ecol. Environ. 2017, 2, 236–249. [Google Scholar] [CrossRef]
- Hobbie, S.E. Plant species effects on nutrient cycling: Revisiting litter feedbacks. Trends Ecol. Evol. 2015, 30, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Hättenschwiler, S.; Tiunov, A.V.; Scheu, S. Biodiversity and litter decomposition in terrestrial ecosystems. Annu. Rev. Ecol. Evo. Syst. 2005, 36, 191–218. [Google Scholar] [CrossRef]
- Simpson, A.J.; Simpson, M.J.; Smith, E.; Kelleher, B.P. Microbially derived inputs to soil organic matter: Are current estimates too low? Environ. Sci. Technol. 2007, 41, 8070–8076. [Google Scholar] [CrossRef]
- Hector, A.; Beale, A.J.; Minns, A.; Otway, S.J.; Lawton, J.H. Consequences of the reduction of plant diversity for litter decomposition: Effects through litter quality and microenvironment. Oikos 2000, 90, 357–371. [Google Scholar] [CrossRef]
- Handa, I.T.; Aerts, R.; Berendse, F.; Berg, M.P.; Bruder, A.; Butenschoen, O.; Chauvet, E.; Gessner, M.O.; Jabiol, J.; Makkonen, M. Consequences of biodiversity loss for litter decomposition across biomes. Nature 2014, 509, 218–221. [Google Scholar] [CrossRef]
- Hu, X.; Arif, M.; Ding, D.; Li, J.; He, X.; Li, C. Invasive Plants and Species Richness Impact Litter Decomposition in Riparian Zones. Front. Plant Sci. 2022, 13, 955656. [Google Scholar] [CrossRef]
- Janusauskaite, D.; Straigyte, L. Leaf litter decomposition differences between alien and native maple species. Balt. For. 2011, 17, 189–196. [Google Scholar]
- Van Kleunen, M.; Weber, E.; Fischer, M. A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol. Lett. 2010, 13, 235–245. [Google Scholar] [CrossRef]
- Zubek, S.; Majewska, M.L.; Blaszkowski, J.; Stefanowicz, A.M.; Nobis, M.; Kapusta, P. Invasive plants affect arbuscular mycorrhizal fungi abundance and species richness as well the performance of native plants grown in invaded soils. Biol. Fertil. Soils. 2016, 52, 879–893. [Google Scholar] [CrossRef]
- Liao, C.; Peng, R.; Luo, Y.; Zhou, X.; Wu, X.; Fang, C.; Chen, J.; Li, B. Altered ecosystem carbon and nitrogen cycles by plant invasion: A meta-analysis. New Phytol. 2008, 177, 706–714. [Google Scholar] [CrossRef]
- McTee, M.R.; Lekberg, Y.; Mummey, D.; Rummel, A.; Ramsey, P.W. Do invasive plants structure microbial communities to accelerate decomposition in intermountain grasslands? Ecol. Evol. 2017, 7, 11227–11235. [Google Scholar] [CrossRef]
- Arthur, M.A.; Bray, S.R.; Kuchle, C.R.; McEwan, R.W. The influence of the invasive shrub, Lonicera maackii, on leaf decomposition and microbial community dynamics. Plant Ecol. 2012, 213, 1571–1582. [Google Scholar] [CrossRef]
- Allison, S.D.; Vitousek, P.M. Rapid nutrient cycling in leaf litter from invasive plants in Hawai’i. Oecologia 2004, 141, 612–619. [Google Scholar] [CrossRef] [PubMed]
- Jo, I.; Fridley, J.D.; Frank, D.A. Invasive plants accelerate nitrogen cycling: Evidence from experimental woody monocultures. J. Ecol. 2017, 105, 1105–1110. [Google Scholar] [CrossRef]
- Cleveland, C.C.; Reed, S.C.; Keller, A.B.; Nemergut, D.R.; O’Neill, S.P.; Ostertag, R.; Vitousek, P.M. Litter quality versus soil microbial community controls over decomposition: A quantitative analysis. Oecologia 2014, 174, 283–294. [Google Scholar] [CrossRef]
- Kourtev, P.S.; Ehrenfeld, J.G.; Haggblom, M. Exotic plant species alter the microbial community structure and function in the soil. Ecology 2002, 83, 3152–3166. [Google Scholar] [CrossRef]
- Kourtev, P.S.; Ehrenfeld, J.G.; Häggblom, M. Experimental analysis of the effect of exotic and native plant species on the structure and function of soil microbial communities. Soil Biol. Biochem. 2003, 35, 895–905. [Google Scholar] [CrossRef]
- Mincheva, T.; Barni, E.; Varese, G.C.; Brusa, G.; Cerabolini, B.; Siniscalco, C. Litter quality, decomposition rates andsaprotrophicmycoflora in Fallopiajaponica (Houtt.) RonseDecraene and in adjacent native grassland vegetation. Acta Oecol. 2014, 54, 29–35. [Google Scholar] [CrossRef]
- Stefanowicz, A.M.; Stanek, M.; Nobis, M.; Zubek, S. Species-specific effects of plant invasions on activity, biomass, and composition of soil microbial communities. Biol. Fertil. Soils. 2016, 52, 841–852. [Google Scholar] [CrossRef]
- Hoyos-Santillan, J.; Lomax, B.H.; Turner, B.L.; Sjögersten, S. Nutrient limitation or home field advantage: Does microbial community adaptation overcome nutrient limitation of litter decomposition in a tropical peatland? J. Ecol. 2018, 106, 1558–1569. [Google Scholar] [CrossRef]
- Kaiser, C.; Franklin, O.; Dieckmann, U.; Richter, A. Microbial community dynamics alleviate stoichiometric constraints during litter decay. Ecol. Lett. 2014, 17, 680–690. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, B.E.; Klironomos, J.N. Breaking new ground: Soil communities and exotic plant invasion. Bioscience 2005, 55, 477–487. [Google Scholar] [CrossRef]
- van der Putten, W.; Klironomos, J.; Wardle, D. Microbial ecology of biological invasions. ISME J. 2007, 1, 28–37. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Chang, S.X.; Liang, X.; Qin, H.; Chen, J.; Xu, Q. Linking soil fungal community structure and function to soil organic carbon chemical composition in intensively managed subtropical bamboo forests. Soil Biol. Biochem. 2017, 107, 19–31. [Google Scholar] [CrossRef]
- Chukwuma, O.B.; Rafatullah, M.; Tajarudin, H.A.; Ismail, N. Lignocellulolytic Enzymes in Biotechnologi-cal and Industrial Processes: A Review. Sustainability 2020, 12, 7282. [Google Scholar] [CrossRef]
- Lombard, V.; GolacondaRamulu, H.; Drula, E.; Coutinho, P.M.; Henrissat, B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014, 42, D490–D495. [Google Scholar] [CrossRef]
- Swift, M.J.; Heal, O.W.; Anderson, J.M. Decomposition in terrestrial ecosystems. Stud. Ecol. 1979, 5, 372. [Google Scholar]
- Vogt, K.A.; Grier, C.C.; Vogt, D.J. Production, turnover, and nutrient dynamics of above- and belowground detritus of world forests. Adv. Ecol. Res. 1986, 15, 303–377. [Google Scholar] [CrossRef]
- Biasi, C.; Fontana, L.E.; Restello, R.M.; Hepp, L.U. Effect of invasive Hovenia dulcis on microbial decomposition and diversity of hyphomycetes in Atlantic forest streams. Fungal Ecol. 2020, 44, 100890. [Google Scholar] [CrossRef]
- McCary, M.A.; Wise, D.H. Plant invader alters soil food web via changes to fungal resources. Oecologia 2019, 191, 587–599. [Google Scholar] [CrossRef] [PubMed]
- Woodworth, G.R.; Ward, J.N.; Carr, D.E. Exotic tree and shrub invasions alter leaf-litter microflora and arthropod communities. Oecologia 2020, 193, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Li, B.; Wu, J.; Hu, S. Invasive plants differentially affect soil biota through litter and rhizosphere pathways: A meta-analysis. Ecol. Lett. 2019, 22, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Chiba de Castro, W.A.; Almeida, R.V.; Xavier, R.O.; Bianchini, I.; Moya, H.; Silva Matos, D.M. Litter accumulation and biomass dynamics in riparian zones in tropical South America of the Asian invasive plant Hedychium coronarium J. König (Zingeraceae). Plant Ecol. Divers. 2019, 13, 47–59. [Google Scholar] [CrossRef]
- Incerti, G.; Carteni, F.; Cesarano, G.; Sarker, T.; El-Gawad, A.M.; D’Ascoli, R.; Bonanomi, G.; Giannino, F. Faster N release, but not C loss, from leaf litter of invasives compared to native species in Mediterranean ecosystems. Front. Plant Sci. 2018, 9, 534. [Google Scholar] [CrossRef]
- Matson, P. Plant-soil interactions in primary succession at Hawaii Volcanoes National Park. Oecologia 1990, 85, 241–246. [Google Scholar] [CrossRef]
- Maule, H.G.; Andrews, M.; Morton, J.D.; Jones, A.V.; Daly, G.T. Sun/shade acclimation and nitrogen nutrition of Tradescantia fluminensis, a problem weed in New Zealand native forest remnants. N. Z. J. Ecol. 1995, 19, 35–46. [Google Scholar]
- Standish, R.J.; Williams, P.A.; Robertson, A.W.; Scott, N.A.; Hedderley, D.I. Invasion by a perennial herb increases decomposition rate and alters nutrient availability in warm temperate lowland forest remnants. Biol. Invasions 2004, 6, 71–81. [Google Scholar] [CrossRef]
- Vinton, M.A.; Goergen, E.M. Plant-soil feedbacks contribute to the persistence of Bromus inermis in tallgrass prairie. Ecosystems 2006, 9, 967–976. [Google Scholar] [CrossRef]
- Suding, K.N.; Collins, S.L.; Gough, L.; Clark, C.; Cleland, E.E.; Gross, K.; Milchunas, D.G.; Pennings, S. Functional- and abundance-based mechanisms explain diversity loss due to N fertilization. Proc. Natl. Acad. Sci. USA 2005, 102, 4387–4392. [Google Scholar] [CrossRef]
- Zhang, D.; Hui, D.; Luo, Y.; Zhou, G. Rates of litter decomposition in terrestrial ecosystems: Global patterns and controlling factors. J. Plant Ecol. 2008, 1, 85–93. [Google Scholar] [CrossRef]
- Songwe, N.; Okali, D.U.; Fasehum, F. Litter decomposition and nutrient release in a tropical rainforest, Southern Bakundu Forest Reserve, Cameroon. J. Trop. Ecol. 1995, 11, 333–350. [Google Scholar] [CrossRef]
- Gessner, M.O.; Swan, C.M.; Dang, C.K.; McKie, B.G.; Bardgett, R.D.; Wall, D.H.; Hättenschwiler, S. Diversity meets decomposition. Trends Ecol. Evol. 2010, 25, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Emery, S.L.; Perry, J.A. Decomposition rates and phosphorus concentrations of purple loosestrife (Lythrumsalicaria) and cattail (Typha spp.) in fourteen Minnesota Wetlands. Hydrobiologia 1996, 323, 129–138. [Google Scholar] [CrossRef]
- Lecerf, A.; Marie, G.; Kominoski, J.S.; LeRoy, C.J.; Bernadet, C.; Swan, C.M. Incubation time, functional litter diversity, and habitat characteristics predict litter-mixing effects on decomposition. Ecology 2011, 92, 160–169. [Google Scholar] [CrossRef]
- Silva, D.S.; Cunha-Santino, M.B.; Marques, E.E.; Bianchini, I., Jr. The decomposition of aquatic macrophytes: Bioassays versus in situ experiments. Hydrobiologia 2011, 665, 219–227. [Google Scholar] [CrossRef]
- Vignati, D.A.L.; Ferrari, B.J.D.; Dominik, J. Laboratory-to-field extrapolation in aquatic sciences: Conceptual frameworks are needed to narrow the gap between laboratory- and field-based research. Environ. Sci. Technol. 2007, 41, 1067–1073. [Google Scholar] [CrossRef]
- Santos, M.G.; Cunha-Santino, M.B.; Bianchini, I., Jr. Photodegradation, chemical and biologic oxidations from mineralization of Utricularia breviscapa leachate. Acta Limnol. Bras. 2006, 18, 347–355. [Google Scholar]
- Nunes, M.F.; Cunha-Santino, M.B.; Bianchini, I., Jr. Aerobic mineralization of carbon and nitrogen from Myriophyllum aquaticum (Vell.) Verdc. leachate. Acta Limnol. Bras. 2007, 19, 285–293. [Google Scholar]
- Bradford, M.A.; Berg, B.; Maynard, D.S.; Wieder, W.R.; Wood, S.A. Understanding the dominant controls on litter decomposition. J. Ecol. 2016, 104, 229–238. [Google Scholar] [CrossRef]
- Makkonen, M.; Berg, M.P.; Handa, I.T.; Hättenschwiler, S.; Ruijven, J.; Bodegom, P.M.; Aerts, R. Highly consistent effects of plant litter identity and functional traits on decomposition across a latitudinal gradient. Ecol. Lett. 2012, 15, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Prescott, C.E. Litter decomposition: What controls it and how can we alter it to sequester more carbon in forest soils? Biogeochemistry 2010, 101, 133–149. [Google Scholar] [CrossRef]
- Smith, V.C.; Bradford, M.A. Litter quality impacts on grassland litter decomposition are differently dependent on soil fauna across times. Appl. Soil Ecol. 2003, 24, 197–203. [Google Scholar] [CrossRef]
- Garcia-Palacios, P.; Maestre, F.T.; Kattge, J.; Wall, D.H. Climate and litter quality differently modulate the effects of soil fauna on litter decomposition across biomes. Ecol. Lett. 2015, 16, 1045–1053. [Google Scholar] [CrossRef]
- Souza, C.N. Diversidade de Fungos do Solo da Mata Atlântica. Master’s Thesis, UFLA, Lavras, Brazil, 2010. p. 66. [Google Scholar]
- Sanches, L.; Valentini, C.M.A.; Biudes, M.S.; Nogueira, J.S. Dinâmica sazonal da produção e decomposição de serrapilheira em floresta tropical de transição. Rev. Bras. Eng. Agr. Amb. 2009, 13, 183–189. [Google Scholar] [CrossRef]
- Boer, W.; Folman, L.B.; Summerbell, R.C.; Boddy, L. Living in a fungal world: Impact of fungi on soil bacterial niche development. FEMS Microbiol. Rev. 2005, 29, 795–811. [Google Scholar] [CrossRef]
- Costa, P.M.O. Dinâmica de Serapilheira e Diversidade de Fungos em Solo de Sistema Agroflorestal. Ph.D. Thesis, Universidade Federal de Pernambuco, Centro de Biociências, Recife, Brazil, 2015. [Google Scholar]
- Marques, M.F.O.; Gusmão, L.F.P.; Maia, L.C. Riqueza de espécies de fungos conidiais em duas áreas de Mata Atlântica no Morro da Pioneira, Serra da Jibóia, BA, Brasil. Acta Bot. Bras. 2008, 22, 954–961. [Google Scholar] [CrossRef]
- Pathak, A.; Jaswal, R.; Xu, X.; White, J.R.; Edwards, B., III; Hunt, J.; Brooks, S.; Rathore, R.S.; Agarwal, M.; Chauhan, A. Characterization of bacterial and fungal assemblages from historically contaminated metalliferous soils using metagenomics coupled with diffusion chambers and microbial traps. Front. Microbiol. 2020, 11, 1024. [Google Scholar] [CrossRef]
- Rahouti, M.; Steiman, R.; Seigle-Murandi, F.; Christov, L.P. Growth of 1044 strains and species of fungi on 7 phenolic lignin model compounds. Chemosphere 1999, 38, 2549–2559. [Google Scholar] [CrossRef]
- Zhang, Y.H.; Hong, J.; Ye, X. Cellulase assays. In Biofuels. Methods in Molecular Biology (Methods and Protocols); Mielenz, J., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 581. [Google Scholar]
- Barrett, K.; Jensen, K.; Meyer, A.S. Fungal secretome profile categorization of CAZymes by function and family corresponds to fungal phylogeny and taxonomy: Example Aspergillus and Penicillium. Sci. Rep. 2020, 10, 5158. [Google Scholar] [CrossRef]
- Egidi, E.; Delgado-Baquerizo, M.; Plett, J.M. A few Ascomycota taxa dominate soil fungal communities worldwide. Nat. Commun. 2019, 10, 2369. [Google Scholar] [CrossRef]
- Moura, D.R.; Araújo, E.C.G.; Silva, T.C.; Leão, S.L.M.; Lima, T.V. Efeitos alelopáticos de extratos de Tradescantia zebrina na germinação de Lactuca sativa. Ecol. Nut. Florest. 2018, 6, 45–50. [Google Scholar] [CrossRef]
- Navarro, G.B.; Pelier, L.C.; Favier, M.M. Efectos alelopáticos de las coberturas vivas Commelina diffusa Burm. F. y Tradescantia zebrina Shunltz sobre Coffea arabica L. Cent. Agric. 2018, 40, 75–78. [Google Scholar]
- Abhijeet, R.; Aiswarya, J.; Raja Rajeswary, T.; Annamalai, A.; Lakshmi, P.T.V. Genome-wide annotation, comparison and functional genomics of carbohydrate-active enzymes in legumes infecting Fusarium oxysporum formae speciales. Mycology 2020, 11, 56–70. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Wang, C.; Xu, J.-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genom. 2013, 14, 274. [Google Scholar] [CrossRef] [PubMed]
- Voříšková, J.; Baldrian, P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 2013, 7, 477–486. [Google Scholar] [CrossRef]
- Frankland, J.C. Fungal succession—Unravelling the unpredictable. Mycol. Res. 1998, 102, 1–15. [Google Scholar] [CrossRef]
- Keller, A.B.; Phillips, R.P. Leaf litter decay rates differ between mycorrhizal groups in temperate, but not tropical, forests. New Phytol. 2019, 222, 556–564. [Google Scholar] [CrossRef]
- Kohlmeier, S.; Smits, T.H.M.; Ford, R.M.; Keel, C.; Harms, H.; Wick, L.Y. Taking the fungal highway, mobilization of pollutant-degrading bacteria by fungi. Environ. Sci. Technol. 2005, 39, 4640–4646. [Google Scholar] [CrossRef]
- Rodolfo, A.M.; Temponi, L.G.; Junior, J.F.C. Levantamento de plantas exóticas na trilha do Poço Preto, Parque Nacional do Iguaçu, Paraná, Brasil. Rev. Bras. Bioc. 2008, 6, 22–24. Available online: https://www.seer.ufrgs.br/index.php/rbrasbioci/article/view/114974 (accessed on 22 November 2021).
- World Wide Fund for Nature Brazil. Descobrindo a Fauna do Parque Nacional do Iguaçu. Available online: https://www.wwf.org.br/informacoes/sala_de_imprensa/?40523/Descobrindo-a-fauna-do-Parque-Nacional-do-Iguau (accessed on 10 March 2020).
- Instituto Ambiental do Paraná (IAP). Portaria iap nº 059, de 15 de Abril de 2015. Available online: http://www.iap.pr.gov.br/arquivos/File/Lista_invasoras_PR_corrigida_set_2015.pdf (accessed on 10 March 2020).
- Mantoani, M.C.; Dias, J.; Orsi, M.L.; Torezan, J.M.D. Efeitos da invasão por Tradescantia zebrina Heynh. sobre regenerantes de plantas arbóreas em um fragmento de floresta estacional semidecidual secundária em Londrina (Pr). Biotemas 2013, 26, 63–70. [Google Scholar] [CrossRef]
- Zenni, R.D.; Ziller, S.R. An overview of invasive plants in Brazil. Rev. Bras. Bot. 2011, 34, 431–446. [Google Scholar] [CrossRef]
- Lorenzi, H.; Souza, H.M. Plantas Ornamentais No Brasil: Arbustivas, Herbáceas e Trepadeiras, 4th ed.; Nova Odessa, Instituto Plantarum: São Paulo, Brazil, 2008; 1120p. [Google Scholar]
- Chiba de Castro, W.A.; Xavier, R.O.; Garrido, F.H.L.; Castro, J.H.C.; Peres, C.K.; Luz, R.C. Fraying around the edges: Negative effects of the invasive Tradescantia zebrina Hort. ex Bosse (Commelinaceae) on tree regeneration in the Atlantic Forest under different competitive and environmental conditions. J. Plant Ecol. 2019, 12, 713–721. [Google Scholar] [CrossRef]
- Sampaio, A.B.; Schimidt, I.B. Espécies Exóticas Invasoras em Unidades de Conservação Federais do Brasil. BioBrasil Biod. Bras. Rev. Cien. 2013, 3, 2. Available online: http://quintalflorestal.com.br/wp-content/uploads/2017/05/Especies-Exoticas-e-Invasoras-em-Unidades-de-Conservacao-Federais-no-Brasilpdf.pdf (accessed on 21 February 2023).
- Ziller, S.R.; Dechoum, M.S. Plantas e vertebrados exóticos invasores em unidades de conservação no Brasil. BioBrasil Biod. Bras. Rev. Cien. 2013, 3, 2. Available online: https://leimac.sites.ufsc.br/wp-content/uploads/2019/05/Ziller-Dechoum-Plantas-e-vertebrados-exóticos-invasores-em-unidades-de-conservação-no-Brasil.pdf (accessed on 21 February 2023).
- CABI Compendium Invasive Species. Cabi Digital Library. 2021. Available online: https://www.cabidigitallibrary.org/product/QI (accessed on 8 April 2022).
- Martins, B.A.; Pastorini, L.H.; Roberto, B.A.C. Extratos foliares de Tradescantia zebrina Heynh. Prejudicam a germinação e crescimento inicial de Lactuca sativa L. e Solanum lycopersicum L. Enciclopédia Biosf. 2014, 10, 1097–1107. Available online: https://conhecer.org.br/ojs/index.php/biosfera/article/view/2340 (accessed on 21 February 2023).
- Silva, A.S.A.; Voltolini, J.C. Impacto e manejo da invasora exótica Tradescantia zebrina Heynh. exBosse (Commelinaceae) sobre plantas nativas em um fragmento de floresta atlântica no sudeste do Brasil. Botânica 2017, 70, 205–212. Available online: http://www.anchietano.unisinos.br/publicacoes/botanica/volumes/070/011.pdf (accessed on 21 February 2023).
- Chiba de Castro, W.A.; Luz, R.C.; Peres, C.K. Seasonality and forest edge as drivers of Tradescantia zebrina Hort. ex Bosse invasion in the Atlantic Forest. Braz. J. Biol. 2022, 82, e238403. [Google Scholar] [CrossRef]
- Castro, J.H.R.; Garrido, F.L.; Luz, R.C.; Peres, C.K.; Duarte, C.F.; Esparza, S.N.; Cobo, K.A.; Chiba de Castro, W.A. Apparent competition of the invasive inch plant in Atlantic forest. In II International Symposium of Ecology—Ecology in the Anthropocene; UFSCar: São Paulo, Brazil, 2016. [Google Scholar]
- Paulus, B.C. The Diversity and Distribution of Microfungi in Leaf Litter of an Australian WET tropics Rainforest. Ph.D. Thesis, James Cook University, Douglas, QLD, Australia, 2004. [Google Scholar]
- Bills, G.F.; Polishook, J.D. Abundance and diversity of microfungi in leaf litter of a lowland rain forest in Costa Rica. Mycologia 1994, 86, 187–198. [Google Scholar] [CrossRef]
- Machado, C.F. Avaliação da Presença de Microrganismos Indicadores de Contaminação e Patogênicos em Líquidos Lixiviados do Aterro Sanitário de Belo Horizonte. Master’s Thesis, Universidade Federal de Minas Gerais, Escola de Engenharia da UFMG, Minas Gerais, Brazil, 2004. [Google Scholar]
- Nicolau, P.B. Métodos em Microbiologia Ambiental. Universidade Aberta (UAB). 2014. Available online: https://repositorioaberto.uab.pt/bitstream/10400.2/6136/1/UT3_metodos_em_micro_ambiental.pdf (accessed on 10 March 2020).
- Kiiskinen, L.L.; Rättö, M.; Kruus, K. Screening for novel laccase-producing microbes. J. Appl. Microbiol. 2004, 97, 640–646. [Google Scholar] [CrossRef]
- Makhuvele, R.; Ncube, I.; Rensburg, E.L.J.; Grange, D.C. Isolation of fungi from dung of wild herbivores for application in bioethanol production. Braz. J. Microbiol. 2017, 48, 648–655. [Google Scholar] [CrossRef]
- Costa, L.A.; Peixoto, P.E.C.; Gusmão, L.P.F. Effects of storage conditions and culture media on the saprobic fungi diversity in tropical leaf litter. Mycosphere 2015, 6, 43–52. [Google Scholar] [CrossRef]
- Castellani, A. Viability of some pathogenic fungi in distilled water. J. Trop. Med. Hyg. 1939, 24, 270–276. [Google Scholar]
- Lacap, D.C.; Hyde, K.D.; Liew, E.C.Y. An evaluation of the fungal “morphotype” concept based on ribosomal DNA sequences. Fungal Divers. 2003, 12, 53–66. [Google Scholar]
- Gaddeyya, G.; Niharika, P.S.; Bharathi, P.; Kumar, P.K.R. Isolation and identification of soil mycoflora in different crop fields at Salur Mandal. Adv. Appl. Sci. Res. 2012, 3, 2020–2026. [Google Scholar]
- Raeder, U.; Broda, P. Rapid preparation of DNA from filamentous fungi. Let. App. Mic. 1985, 1, 17–20. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols, a Guide to Methods and Applications; Innis, A.M., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 70 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef]
- Olson, J.S. Energy storage and the balance of producers and decomposers in ecological systems. Ecology 1963, 44, 322–331. [Google Scholar] [CrossRef]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting Linear Mixed-Effects Models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- Crawley, M.J. Statistics: An Introduction Using R; Wiley: London, UK, 2005. [Google Scholar]
- Jaccard, P. The Distribution of the Flora of the Alpine Zone. New Phytol. 1912, 11, 37–50. Available online: http://www.jstor.org/stable/2427226 (accessed on 21 February 2023). [CrossRef]
- Baselga, A. Partitioning the turnover and nestedness components of beta diversity. Glob. Ecol. Biogeogr. 2010, 19, 134–143. [Google Scholar] [CrossRef]
- Baselga, A.; Orme, C.D.L. Betapart: An R package for the study of beta diversity. Met. Ecol. Evol. 2012, 3, 808–812. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2008, 26, 32–46. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Henry, M.; Stevens, H.; Szoecs, E.; et al. Vegan: Community Ecology Package. R Package Version 2.3-1. 2019. Available online: https://cran.r-project.org/web/packages/vegan/vegan.pdf (accessed on 3 June 2020).
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: https://www.R-project.org/ (accessed on 10 March 2020).



| Experiment | Litter Bags | Treatment | r2 | k (Days−1) | k (SD) | t½ |
|---|---|---|---|---|---|---|
| In situ | T. zebrine | invaded | 0.81 | 0.01165 | 0.00251 | 59 |
| non-invaded | 0.89 | 0.01316 | 0.00227 | 53 | ||
| Natives | invaded | 0.83 | 0.00550 | 0.00096 | 126 | |
| non-invaded | 0.76 | 0.00519 | 0.00110 | 134 | ||
| In vitro | T. zebrine | invaded | 0.79 | 0.00302 | 0.00055 | 230 |
| non-invaded | 0.89 | 0.00327 | 0.00042 | 212 | ||
| Natives | invaded | 0.73 | 0.00220 | 0.00045 | 315 | |
| non-invaded | 0.81 | 0.00219 | 0.00037 | 317 |
| Response Variable | Factor | Estimate | SE | DF | T Value | p Value |
|---|---|---|---|---|---|---|
| T. zebrina litter | In situ | −0.01647 | 0.00986 | 80 | −1.670 | 0.099 |
| In vitro | −0.001068 | 0.019613 | 44 | −0.054 | 0.957 | |
| Native litter | In situ | −0.009728 | 0.009725 | 80 | −1.00 | 0.320 |
| In vitro | 0.004726 | 0.015915 | 44 | 0.297 | 0.768 |
| Factor | df | Sum of Squares | Mean Squares | F Value | p Value |
|---|---|---|---|---|---|
| Time | 1 | 1.756 | 1.756 | 4.313 | *** |
| Environment | 1 | 0.417 | 0.417 | 0.024 | 0.388 |
| Litter | 1 | 0.382 | 0.382 | 0.022 | 0.563 |
| Time × Environment | 1 | 0.3871 | 0.38707 | 0.9509 | 0.53095 |
| Time × Litter | 1 | 0.4101 | 0.41009 | 1.0075 | 0.41256 |
| Environment × Litter | 1 | 0.5309 | 0.53094 | 1.3044 | 0.08549 |
| Time × Environment × Litter | 1 | 0.4363 | 0.43630 | 1.0719 | 0.30807 |
| Residuals | 32 | 13.0254 | 0.40704 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiba de Castro, W.A.; Vaz, G.C.d.O.; Silva Matos, D.M.d.; Vale, A.H.; Bueno, A.C.P.; Fagundes, L.F.G.; Costa, L.d.; Bonugli Santos, R.C. The Invasive Tradescantia zebrina Affects Litter Decomposition, but It Does Not Change the Lignocellulolytic Fungal Community in the Atlantic Forest, Brazil. Plants 2023, 12, 2162. https://doi.org/10.3390/plants12112162
Chiba de Castro WA, Vaz GCdO, Silva Matos DMd, Vale AH, Bueno ACP, Fagundes LFG, Costa Ld, Bonugli Santos RC. The Invasive Tradescantia zebrina Affects Litter Decomposition, but It Does Not Change the Lignocellulolytic Fungal Community in the Atlantic Forest, Brazil. Plants. 2023; 12(11):2162. https://doi.org/10.3390/plants12112162
Chicago/Turabian StyleChiba de Castro, Wagner Antonio, Giselle Cristina de Oliveira Vaz, Dalva Maria da Silva Matos, Alvaro Herrera Vale, Any Caroline Pantaleão Bueno, Luiz Fernando Grandi Fagundes, Letícia da Costa, and Rafaella Costa Bonugli Santos. 2023. "The Invasive Tradescantia zebrina Affects Litter Decomposition, but It Does Not Change the Lignocellulolytic Fungal Community in the Atlantic Forest, Brazil" Plants 12, no. 11: 2162. https://doi.org/10.3390/plants12112162
APA StyleChiba de Castro, W. A., Vaz, G. C. d. O., Silva Matos, D. M. d., Vale, A. H., Bueno, A. C. P., Fagundes, L. F. G., Costa, L. d., & Bonugli Santos, R. C. (2023). The Invasive Tradescantia zebrina Affects Litter Decomposition, but It Does Not Change the Lignocellulolytic Fungal Community in the Atlantic Forest, Brazil. Plants, 12(11), 2162. https://doi.org/10.3390/plants12112162