Identification of Candidate Genes Involved in Fruit Ripening and Crispness Retention Through Transcriptome Analyses of a ‘Honeycrisp’ Population
Abstract
:1. Introduction
2. Results
2.1. Phenotype and Transcriptome Variations Among the Individuals
2.2. Functional Annotation of Differentially Expressed Genes
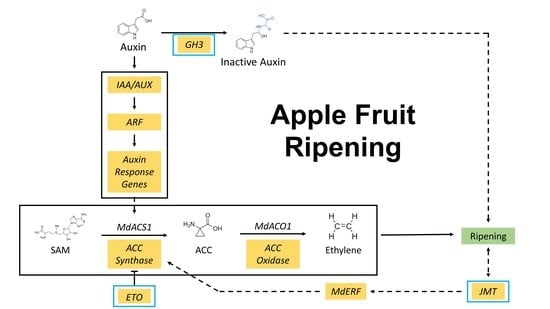
2.3. The Expression Patterns of Auxin- and Ethylene-Related Genes
2.4. The Expression Patterns of Cell Wall-Related Genes
2.5. RNA-Seq Results Validation Using nCounter® Technology and qRT-PCR
3. Discussion
3.1. Fruit Ripening and Crispness Retention
3.2. Cell Wall-Related Genes and Crispness Retention
4. Conclusions
5. Materials and Methods
5.1. Plant Materials
5.2. RNA Sample Preparation and RNA Sequencing
5.3. Differential Expression Analysis
5.4. Gene Validation Using NanoString nCounter® and qRT-PCR
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Data Availability
References
- Luby, J.J.; Bedford, D.S. rep. Honeycrisp apple. In Minnesota Report; Minnesota Agricultural Experiment Station: Saint Paul, MN, USA, 1992. [Google Scholar]
- Washington Tree Fruit Acreage Report. Available online: https://www.nass.usda.gov/Statistics_by_State/Washington/Publications/Fruit/2017/FT2017.pdf (accessed on 12 August 2020).
- Hampson, C.; Quamme, H.; Hall, J. Using Sensory Evaluation Panels to Screen Apple Breeding Selections. Acta Horticulturae 2000, 538, 201–205. [Google Scholar] [CrossRef]
- Péneau, S.; Hoehn, E.; Roth, H.R.; Escher, F.; Nuessli, J. Importance and Consumer Perception of Freshness of Apples. Food Qual. Prefer. 2006, 17, 9–19. [Google Scholar] [CrossRef]
- Tong, C.; Krueger, D.; Vickers, Z.; Bedford, D.; Luby, J.; El-Shiekh, A.; Shackel, K.; Ahmadi, H. Comparison of Softening-Related Changes during Storage of ’Honeycrisp’ Apple, Its Parents, and ’Delicious’. J. Am. Soc. Hort. Sci. 1999, 124, 407–415. [Google Scholar] [CrossRef] [Green Version]
- Barry, C.S.; Giovannoni, J.J. Ethylene and Fruit Ripening. J. Plant Growth Regul. 2007, 26, 143–159. [Google Scholar] [CrossRef]
- Johnston, J.W.; Hewett, E.W.; Hertog, M.L. Postharvest Softening of Apple (Malus Domestica) Fruit: A Review. N. Z. J. Crop Hort. Sci. 2002, 30, 145–160. [Google Scholar] [CrossRef]
- Harb, J.; Gapper, N.E.; Giovannoni, J.J.; Watkins, C.B. Molecular Analysis of Softening and Ethylene Synthesis and Signaling Pathways in a Non-Softening Apple Cultivar, ’Honeycrisp’ and a Rapidly Softening Cultivar, ’McIntosh’. Postharvest Biol. Technol. 2012, 64, 94–103. [Google Scholar] [CrossRef]
- Costa, F.; Stella, S.; Weg, W.E.V.D.; Guerra, W.; Cecchinel, M.; Dallavia, J.; Koller, B.; Sansavini, S. Role of the Genes Md-ACO1 and Md-ACS1 in Ethylene Production and Shelf Life of Apple (Malus Domestica Borkh). Euphytica 2005, 141, 181–190. [Google Scholar] [CrossRef]
- Costa, F.; Peace, C.P.; Stella, S.; Serra, S.; Musacchi, S.; Bazzani, M.; Sansavini, S.; Weg, W.E.V.D. QTL Dynamics for Fruit Firmness and Softening around an Ethylene-Dependent Polygalacturonase Gene in Apple (Malusxdomestica Borkh.). J. Exp. Bot. 2010, 61, 3029–3039. [Google Scholar] [CrossRef] [Green Version]
- Harada, T.; Sunako, T.; Wakasa, Y.; Soejima, J.; Satoh, T.; Niizeki, M. An Allele of the 1-Aminocyclopropane-1-Carboxylate Synthase Gene (Md-ACS1) Accounts for the Low Level of Ethylene Production in Climacteric Fruits of Some Apple Cultivars. Theor. Appl. Genet. 2000, 101, 742–746. [Google Scholar] [CrossRef]
- Dougherty, L.; Zhu, Y.; Xu, K. Assessing the Allelotypic Effect of Two Aminocyclopropane Carboxylic Acid Synthase-Encoding Genes MdACS1 and MdACS3a on Fruit Ethylene Production and Softening in Malus. Hort. Res. 2016, 3, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Harker, F.; Stec, M.G.; Hallett, I.C.; Bennett, C.L. Texture of Parenchymatous Plant Tissue: a Comparison between Tensile and Other Instrumental and Sensory Measurements of Tissue Strength and Juiciness. Postharvest Biol. Technol. 1997, 11, 63–72. [Google Scholar] [CrossRef]
- Wakabayashi, K. Changes in Cell Wall Polysaccharides During Fruit Ripening. J. Plant Sci. 2000, 113, 231–237. [Google Scholar] [CrossRef]
- Caffall, K.H.; Mohnen, D. The Structure, Function, and Biosynthesis of Plant Cell Wall Pectic Polysaccharides. Carbohydr. Res. 2009, 344, 1879–1900. [Google Scholar] [CrossRef] [PubMed]
- Wakasa, Y.; Kudo, H.; Ishikawa, R.; Akada, S.; Senda, M.; Niizeki, M.; Harada, T. Low Expression of an Endopolygalacturonase Gene in Apple Fruit with Long-Term Storage Potential. Postharvest Biol. Technol. 2006, 39, 193–198. [Google Scholar] [CrossRef]
- Goulao, L.; Oliveira, C. Cell Wall Modifications during Fruit Ripening: When a Fruit Is Not the Fruit. Trends Food Sci. Technol 2008, 19, 4–25. [Google Scholar] [CrossRef] [Green Version]
- Nobile, P.M.; Wattebled, F.; Quecini, V.; Girardi, C.L.; Lormeau, M.; Laurens, F. Identification of a Novel α-L-Arabinofuranosidase Gene Associated with Mealiness in Apple. J. Exp. Bot. 2011, 62, 4309–4321. [Google Scholar] [CrossRef] [Green Version]
- Gwanpua, S.G.; Mellidou, I.; Boeckx, J.; Kyomugasho, C.; Bessemans, N.; Verlinden, B.E.; Hertog, M.L.; Hendrickx, M.; Nicolai, B.M.; Geeraerd, A.H. Expression Analysis of Candidate Cell Wall-Related Genes Associated with Changes in Pectin Biochemistry during Postharvest Apple Softening. Postharvest Biol. Technol. 2016, 112, 176–185. [Google Scholar] [CrossRef]
- Gwanpua, S.; Verlinden, B.; Hertog, M.; Nicolaï, B.; Hendrickx, M.; Geeraerd, A. Understanding the Regulation of Texture Degradation during Apple Softening—A Kinetic Modelling Approach. Acta Horticulturae 2018, 1194, 1399–1406. [Google Scholar] [CrossRef]
- Mann, H.S.; Alton, J.J.; Kim, S.; Tong, C.B. Differential Expression of Cell-Wall–Modifying Genes and Novel CDNAs in Apple Fruit During Storage. J. Am. Soc. Hortic. Sci. 2008, 133, 152–157. [Google Scholar] [CrossRef] [Green Version]
- Trujillo, D. Molecular and enzymatic examination of cell wall-modifying proteins in relation to apple crispness maintenance. Master’s Thesis, University of Minnesota, Minneapolis, MN, USA, September 2012. [Google Scholar]
- Atkinson, R.G.; Johnston, S.L.; Yauk, Y.K.; Sharma, N.N.; Schröder, R. Analysis of Xyloglucan Endotransglucosylase/Hydrolase (XTH) Gene Families in Kiwifruit and Apple. Postharvest Biol. Technol. 2009, 51, 149–157. [Google Scholar] [CrossRef]
- Muñoz-Bertomeu, J.; Miedes, E.; Lorences, E. Expression of Xyloglucan Endotransglucosylase/Hydrolase (XTH) Genes and XET Activity in Ethylene Treated Apple and Tomato Fruits. J. Plant Physiol. 2013, 170, 1194–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ireland, H.S.; Gunaseelan, K.; Muddumage, R.; Tacken, E.J.; Putterill, J.; Johnston, J.W.; Schaffer, R.J. Ethylene Regulates Apple (Malus × Domestica) Fruit Softening Through a Dose × Time-Dependent Mechanism and Through Differential Sensitivities and Dependencies of Cell Wall-Modifying Genes. Plant Cell Physiol. 2014, 55, 1005–1016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trujillo, D.I.; Mann, H.S.; Tong, C.B.S. Examination of Expansin Genes as Related to Apple Fruit Crispness. Tree Genet. Genomes 2011, 8, 27–38. [Google Scholar] [CrossRef]
- Chang, H.Y.; Vickers, Z.M.; Tong, C.B.S. The Use of a Combination of Instrumental Methods to Assess Change in Sensory Crispness during Storage of a “Honeycrisp” Apple Breeding Family. J. Texture Stud. 2018, 49, 228–239. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A Revolutionary Tool for Transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, M.M. Digital Multiplexed Gene Expression Analysis Using the NanoString NCounter System. Curr. Protoc. Mol. Biol. 2011, 94, 25B.10.1–25B.10.17. [Google Scholar]
- Kende, H. Ethylene Biosynthesis. Annu. Rev. Plant Biol. 1993, 44, 283–307. [Google Scholar] [CrossRef]
- Tan, D.; Li, T.; Wang, A. Apple 1-Aminocyclopropane-1-Carboxylic Acid Synthase Genes, MdACS1 and MdACS3a, Are Expressed in Different Systems of Ethylene Biosynthesis. Plant Mol. Biol. Rep. 2012, 31, 204–209. [Google Scholar] [CrossRef]
- An, J.P.; Yao, J.F.; Xu, R.R.; You, C.X.; Wang, X.F.; Hao, Y.J. An Apple NAC Transcription Factor Enhances Salt Stress Tolerance by Modulating the Ethylene Response. Physiol. Plant 2018, 164, 279–289. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Quan, P.; Liu, H.; Li, L.; Qi, S.; Zhang, M. Transcriptomic and Metabolic Analyses Provide New Insights into the Apple Fruit Quality Decline during Long-Term Cold Storage. J. Agric. Food Chem. 2020, 68, 4699–4716. [Google Scholar] [CrossRef]
- Yue, P.; Lu, Q.; Liu, Z.; Lv, T.; Li, X.; Bu, H.; Liu, W.; Xu, Y.; Yuan, H.; Wang, A. Auxin-Activated MdARF5 Induces the Expression of Ethylene Biosynthetic Genes to Initiate Apple Fruit Ripening. New Phytol. 2020, 226, 1781–1795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaffer, R.J.; Ireland, H.S.; Ross, J.J.; Ling, T.J.; David, K.M. SEPALLATA1/2-Suppressed Mature Apples Have Low Ethylene, High Auxin and Reduced Transcription of Ripening-Related Genes. AoB PLANTS 2013, 5, pls047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busatto, N.; Tadiello, A.; Trainotti, L.; Costa, F. Climacteric Ripening of Apple Fruit Is Regulated by Transcriptional Circuits Stimulated by Cross-Talks between Ethylene and Auxin. Plant Signal. Behav. 2016, 12, e1268312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, S.; Lee, J.; Rudell, D.; Evans, K.; Zhu, Y. Transcriptional Regulation of Auxin Metabolism and Ethylene Biosynthesis Activation During Apple (Malus × Domestica) Fruit Maturation. J. Plant Growth Regul. 2016, 35, 655–666. [Google Scholar] [CrossRef]
- Devoghalaere, F.; Doucen, T.; Guitton, B.; Keeling, J.; Payne, W.; Ling, T.; Ross, J.; Hallett, I.; Gunaseelan, K.; Dayatilake, G.; et al. A Genomics Approach to Understanding the Role of Auxin in Apple (Malus × Domestica) Fruit Size Control. BMC Plant Biol. 2012, 12, 7. [Google Scholar] [CrossRef] [Green Version]
- Bottcher, C.; Keyzers, R.A.; Boss, P.K.; Davies, C. Sequestration of Auxin by the Indole-3-Acetic Acid-Amido Synthetase GH3-1 in Grape Berry (Vitis Vinifera L.) and the Proposed Role of Auxin Conjugation during Ripening. J. Exp. Bot. 2010, 61, 3615–3625. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Kang, B.C.; Jiang, H.; Moore, S.L.; Li, H.; Watkins, C.B.; Setter, T.L.; Jahn, M.M. A GH3-Like Gene, CcGH3, Isolated from Capsicum Chinense L. Fruit Is Regulated by Auxin and Ethylene. Plant Mol. Biol. Rep. 2005, 58, 447–464. [Google Scholar] [CrossRef]
- Shi, Y.; Jiang, L.; Zhang, L.; Kang, R.; Yu, Z. Erratum: Dynamic Changes in Proteins during Apple (Malus × Domestica) Fruit Ripening and Storage. Hort. Res. 2014, 1, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Song, J.; Campbell-Palmer, L.; Thompson, K.; Li, L.; Walker, B.; Cui, Y.; Li, X. A Proteomic Investigation of Apple Fruit during Ripening and in Response to Ethylene Treatment. J. Proteomics 2013, 93, 276–294. [Google Scholar] [CrossRef]
- Leubner-Metzger, G.; Meins, F. Functions and Regulation of Plant β-(PR-2). In Pathogenesis-related proteins in plants; Datta, S.K., Muthukrishnan, S., Eds.; CRC Press: Boca Raton, FL, USA, 1999; pp. 58–85. [Google Scholar]
- Bai, S.; Dong, C.; Li, B.; Dai, H. A PR-4 Gene Identified from Malus Domestica Is Involved in the Defense Responses against Botryosphaeria Dothidea. Plant Physiol. Biochem. 2013, 62, 23–32. [Google Scholar] [CrossRef]
- Lv, J.; Zhang, M.; Zhang, J.; Ge, Y.; Li, C.; Meng, K.; Li, J. Effects of Methyl Jasmonate on Expression of Genes Involved in Ethylene Biosynthesis and Signaling Pathway during Postharvest Ripening of Apple Fruit. Sci. Hortic. 2018, 229, 157–166. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.; Zhang, L.; Ji, Y.; Tan, D.; Yuan, H.; Wang, A. The Jasmonate-Activated Transcription Factor MdMYC2 Regulates ETHYLENE RESPONSE FACTOR and Ethylene Biosynthetic Genes to Promote Ethylene Biosynthesis during Apple Fruit Ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chae, H.S.; Kieber, J.J. Eto Brute? Role of ACS Turnover in Regulating Ethylene Biosynthesis. Trends Plant Sci. 2005, 10, 291–296. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, H.; Nagata, M.; Saito, K.; Wang, K.L.; Ecker, J.R. Arabidopsis ETO1 specifically interacts with and negatively regulates type 2 1-aminocyclopropane-1-carboxylate synthases. BMC Plant Biol. 2005, 5, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, H.; Wang, Y.; Qi, A.; Zhang, Y.; Xu, J.; Wang, A.; Zhang, Y. PpACS1b, a Pear Gene Encoding ACC Synthase, Is Regulated during Fruit Late Development and Involved in Response to Salicylic Acid. Sci. Hort. 2013, 164, 602–609. [Google Scholar] [CrossRef]
- Wang, K.L.C.; Yoshida, H.; Lurin, C.; Ecker, J.R. Regulation of Ethylene Gas Biosynthesis by the Arabidopsis ETO1 Protein. Nature 2004, 428, 945–950. [Google Scholar] [CrossRef]
- Delmer, D.P.; Amor, Y. Cellulose Biosynthesis. Plant Cell 1995, 7, 987–1000. [Google Scholar]
- Pauly, M.; Gille, S.; Liu, L.; Mansoori, N.; Souza, A.D.; Schultink, A.; Xiong, G. Hemicellulose Biosynthesis. Planta 2013, 238, 627–642. [Google Scholar] [CrossRef]
- Bartley, I.M. Changes in the Glucans of Ripening Apples. Phytochemistry 1976, 15, 625–626. [Google Scholar] [CrossRef]
- Win, N.M.; Yoo, J.; Kwon, S.I.; Watkins, C.B.; Kang, I.K. Characterization of Fruit Quality Attributes and Cell Wall Metabolism in 1-Methylcyclopropene (1-MCP)-Treated ’Summer King’ and ’Green Ball’ Apples During Cold Storage. Front. Plant Sci. 2019, 10, 1513. [Google Scholar] [CrossRef] [Green Version]
- Dheilly, E.; Gall, S.L.; Guillou, M.C.; Renou, J.P.; Bonnin, E.; Orsel, M.; Lahaye, M. Cell Wall Dynamics during Apple Development and Storage Involves Hemicellulose Modifications and Related Expressed Genes. BMC Plant Biol. 2016, 16, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brummell, D.A. Cell Wall Disassembly in Ripening Fruit. Funct. Plant Biol. 2006, 33, 103–119. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, R.G.; Schröder, R.; Hallett, I.C.; Cohen, D.; MacRae, E.A. Overexpression of Polygalacturonase in Transgenic Apple Trees Leads to a Range of Novel Phenotypes Involving Changes in Cell Adhesion. Plant Physiol. 2002, 129, 122–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brummell, D.A.; Harpster, M.H. Cell Wall Metabolism in Fruit Softening and Quality and Its Manipulation in Transgenic Plants. Plant Cell Walls 2001, 47, 311–340. [Google Scholar]
- Yoshioka, H.; Kashimura, Y.; Kaneko, K. β-D-Galactosidase and α-L-Arabinofuranosidase Activities during the Softening of Apples. J. Japan. Soc. Hort. Sci. 1995, 63, 871–878. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Liu, J.; Dang, M.; Zhang, B.; Li, H.; Meng, R.; Qu, D.; Yang, Y.; Zhao, Z. Analysis of β-Galactosidase During Fruit Development and Ripening in Two Different Texture Types of Apple Cultivars. Front. Plant Sci. 2018, 9, 539. [Google Scholar] [CrossRef]
- Hayashi, T. Xyloglucans in the Primary Cell Wall. Annu. Rev. Plant Biol. 1989, 40, 139–168. [Google Scholar]
- Rose, J.K.C.; Braam, J.; Fry, S.C.; Nishitani, K. The XTH Family of Enzymes Involved in Xyloglucan Endotransglucosylation and Endohydrolysis: Current Perspectives and a New Unifying Nomenclature. Plant Cell Physiol. 2002, 43, 1421–1435. [Google Scholar] [CrossRef] [Green Version]
- Miedes, E.; Lorences, E.P. Apple (Malus Domestica) and Tomato (Lycopersicum Esculentum) Fruits Cell-Wall Hemicelluloses and Xyloglucan Degradation DuringPenicillium ExpansumInfection. J. Agric. Food Chem. 2004, 52, 7957–7963. [Google Scholar] [CrossRef]
- Busatto, N.; Farneti, B.; Tadiello, A.; Velasco, R.; Costa, G.; Costa, F. Candidate Gene Expression Profiling Reveals a Time Specific Activation among Different Harvesting Dates in ’Golden Delicious’ and ’Fuji’ Apple Cultivars. Euphytica 2015, 208, 401–413. [Google Scholar] [CrossRef]
- Schmitz, C.A. Enabling Marker-Assisted Breeding for Fruit Texture Traits in Progeny of the Apple Cultivar Honeycrisp. Master’s Thesis, University of Minnesota, Minneapolis, MN, USA, July 2013. [Google Scholar]
- Miedes, E.; Herbers, K.; Sonnewald, U.; Lorences, E.P. Overexpression of a Cell Wall Enzyme Reduces Xyloglucan Depolymerization and Softening of Transgenic Tomato Fruits. J. Agric. Food Chem. 2010, 58, 5708–5713. [Google Scholar] [CrossRef] [PubMed]
- Blanpied, G.D.; Silsby, K.J. Predicting Harvest Date Windows for Apples. Cornell Cooperative Extension 1992, 7, 2–12. [Google Scholar]
- Costa, F.; Cappellin, L.; Longhi, S.; Guerra, W.; Magnago, P.; Porro, D.; Soukoulis, C.; Salvi, S.; Velasco, R.; Biasioli, F.; et al. Assessment of Apple (Malus×Domestica Borkh.) Fruit Texture by a Combined Acoustic-Mechanical Profiling Strategy. Postharvest Biol. Technol. 2011, 61, 21–28. [Google Scholar] [CrossRef]
- López-Gómez, R.; Gómez-Lim, M.A. A Method for Extracting Intact RNA from Fruits Rich in Polysaccharides Using Ripe Mango Mesocarp. HortScience 1992, 27, 440–442. [Google Scholar]
- Andrews, S. FastQC: A quality control tool for high throughput sequence data. Available online: https://userinfo.surfsara.nl/systems/lisa/software/fastqc (accessed on 12 August 2020).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daccord, N.; Celton, J.M.; Linsmith, G.; Becker, C.; Choisne, N.; Schijlen, E.; Geest, H.V.D.; Bianco, L.; Micheletti, D.; Velasco, R.; et al. High-Quality De Novo Assembly of the Apple Genome and Methylome Dynamics of Early Fruit Development. Nat. Genet. 2017, 49, 1099–1106. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Smyth, G.K.; Ritchie, M.; Thorne, N.; Wettenhall, J. LIMMA: Linear Models for Microarray Data. In Bioinformatics and computational biology solutions using R and bioconductor; Gentleman, R., Carey, V.J., Irizarry, R.A., Dudoit, S., Huber, W., Eds.; Springer: New York, NY, USA, 2006; pp. 397–420. [Google Scholar]
- Kolde, R.; Kolde, M. R. Package ’pheatmap’. R package version 1.0.12. Available online: https://CRAN.R-project.org/package=pheatmap (accessed on 12 August 2020).
- Koskinen, P.; Törönen, P.; Nokso-Koivisto, J.; Holm, L. PANNZER: High-Throughput Functional Annotation of Uncharacterized Proteins in an Error-Prone Environment. Bioinformatics 2015, 31, 1544–1552. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene Ontology Analysis for RNA-Seq: Accounting for Selection Bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perini, P.; Pasquali, G.; Margis-Pinheiro, M.; Oliviera, P.R.D.D.; Revers, L.F. Reference Genes for Transcriptional Analysis of Flowering and Fruit Ripening Stages in Apple (Malus × Domestica Borkh.). Mol. Breed. 2014, 34, 829–842. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]






| (a) | At harvest | After Storage | ||
| Retain | Non-crisp | Retain | Non-crisp | |
| α-Arabinoglucosidase | - | MD08G1221800 MD16G1158300 | - | - |
| β-Galactosidase | MD08G1023600 MD15G1251100 | MD00G1018700 MD02G1079200 | MD15G1251100 | - |
| Cellulose synthase/Cellulose synthase- like | MD01G1236500 MD01G1236600 MD02G1311600 MD03G1133700 MD07G1309200 MD08G1147200 MD10G1029800 MD15G1123200 MD15G1340200 MD17G1099800 | MD03G1028900 MD03G1178600 MD13G1209200 MD15G1150300 | MD01G1236500 MD01G1236600 MD08G1076900 MD08G1126200 MD09G1037900 MD15G1064400 MD15G1415100 MD15G1415200 MD17G1144800 | MD03G1028900 MD08G1147200 |
| Expansin | MD00G1125400 MD01G1166700 MD04G1129800 MD07G1233100 MD09G1279500 | MD03G1090700 MD05G1130300 MD06G1041000 MD06G1195100 | MD00G1125400 MD01G1166700 MD04G1052600 | - |
| Galacturonosyltransferase | MD04G1181600 MD05G1363900 MD09G1041100 MD09G1061900 MD11G1318000 MD13G1084900 MD17G1141200 MD16G1084000 | - | MD09G1041100 MD10G1140000 MD13G1084900 MD16G1084000 MD17G1141200 | - |
| Pectate lyase | MD14G1167100 | - | - | MD01G1100600 |
| Pectin methylesterase | MD00G1105300 MD04G1198000 MD07G1289200 MD12G1198000 MD15G1222000 | MD08G1195600 | MD02G1104600 MD13G1149800 MD15G1222000 | MD06G1064700 MD07G1255000 MD08G1195600 MD09G1054900 MD16G1150200 |
| Polygalacturonase | MD01G1068900 MD01G1069000 MD06G1105300 MD09G1290500 MD12G1064100 MD16G1161800 | MD00G1140300 MD03G1162500 MD07G1279000 MD09G1030100 MD09G1030200 MD10G1179100 | MD00G1140300 MD06G1105300 | - |
| Xyloglucan endotransglucosylase/ hydrolase | MD02G1192600 MD09G1102600 MD10G1315100 MD15G1303500 MD16G1014000 MD16G1091200 | MD16G1145200 MD17G1140000 | MD04G1020100 MD15G1303500 MD16G1091200 | MD13G1237300 MD16G1278900 |
| (b) | At harvest | After Storage | ||
| Retain | Lose | Retain | Lose | |
| α-Arabinoglucosidase | - | - | - | MD16G1158300 |
| β-Galactosidase | - | MD00G1018700 MD02G1079200 | - | - |
| Cellulose synthase/ Cellulose synthase- like | MD05G1296600 MD13G1209200 | MD03G1028900 MD15G1415100 MD15G1415200 MD16G1145200 MD17G1038900 | MD05G1296600 MD13G1209200 | MD03G1028900 MD17G1099500 MD17G1099600 |
| Expansin | MD05G1130300 | MD11G1054500 MD16G1070600 | MD05G1130300 MD17G1271500 | MD01G1166700 MD06G1195100 MD07G1233100 MD13G1070200 |
| Galacturonosyltransferase | - | - | MD17G1141200 | - |
| Pectate lyase | - | - | - | MD06G1161400 |
| Pectin methylesterase | MD06G1191000 | MD13G1149800 | - | MD08G1195600 |
| Polygalacturonase | MD06G1105300 | MD10G1179100 | MD13G1092000 | MD00G1087900 MD09G1030100 MD09G1030200 |
| Xyloglucan endotransglucosylase/ hydrolase | MD10G1315100 MD13G1237300 MD15G1303500 MD16G1091200 | MD16G1145200 | MD15G1303500 MD16G1091200 | MD13G1268900 MD16G1145200 |
| (c) | At harvest | After Storage | ||
| Honeycrisp | MN1764 | Honeycrisp | MN1764 | |
| α-Arabinoglucosidase | - | - | - | MD08G1221800 MD16G1158300 |
| β-Galactosidase | - | - | MD08G1139000 MD09G1192500 | MD11G1133400 |
| Cellulose synthase/ Cellulose synthase- like | MD03G1029100 MD03G1178600 MD04G1173700 MD15G1340200 | MD03G1029000 MD03G1133700 MD15G1415100 MD15G1415200 | MD03G1029100 MD03G1178600 MD04G1173700 MD11G1156200 MD13G1209200 MD15G1340200 MD17G1099600 | MD03G1133700 |
| Expansin | - | MD16G1070600 | - | MD04G1052600 MD10G1133200 |
| Galacturonosyltransferase | MD10G1140000 MD17G1141200 | MD00G1136600 MD04G1181600 | MD09G1093100 MD10G1140000 MD11G1318000 | - |
| Pectate lyase | - | MD05G1179500 | - | - |
| Pectin methylesterase | MD01G1220700 MD06G1191000 MD09G1172600 | MD06G1191000 MD08G1195600 MD11G1307500 | MD01G1220700 MD09G1172600 | MD11G1307500 MD16G1150200 |
| Polygalacturonase | MD03G1292400 MD15G1441700 | - | - | MD07G1279000 MD10G1179100 |
| Xyloglucan endotransglucosylase/ hydrolase | MD10G1315100 MD16G1091200 | - | MD16G1091200 | MD09G1152600 MD09G1152700 MD13G1237300 MD17G1140000 |
| (a) Gene ID | At harvest | After storage | At harvest | After storage | Gene function | ||||||||
| HC | MN | Diff. 1 | HC | MN | Diff. | Retain | Lose | Diff. | Retain | Lose | Diff. | ||
| Primary candidate gene | |||||||||||||
| MD01G1062800 | 6052 | 5851 | 0.0 NS | 6731 | 2135 | −1.7 ** | 9720 | 5414 | −0.8 ** | 4488 | 5713 | 0.3 NS | PIP1 | aquaporin |
| MD03G1019900 | 14 | 3 | −2.1 * | 9 | 3 | −1.5 * | 27 | 13 | −1.1 ** | 11 | 6 | −0.8 ** | RLK1 | receptor-like protein kinase |
| MD05G1092300 | 128 | 24 | −2.4 * | 22 | 6 | −1.9 NS | 101 | 9 | −3.4 ** | 12 | 8 | −0.6 NS | GH3 | auxin-responsive protein |
| MD07G1237500 | 659 | 44 | −3.9 ** | 909 | 12 | −6.3 ** | 600 | 253 | −1.2 ** | 269 | 149 | −0.9 ** | RPL | ribosomal protein |
| MD07G1247100 | 561 | 179 | −1.7 ** | 545 | 154 | −1.8 ** | 448 | 274 | −0.7 ** | 377 | 293 | −0.4 * | PMSR | peptide met S-oxide reductase |
| MD07G1259200 | 132 | 3 | −5.3 ** | 138 | 15 | −3.2 ** | 141 | 62 | −1.2 ** | 145 | 84 | −0.8 ** | RPM | disease resistance protein |
| MD07G1270800 | 190 | 27 | −2.8 * | 40 | 23 | −0.8 NS | 209 | 129 | −0.7 ** | 8 | 11 | 0.4 NS | TUB | tubulin |
| MD07G1274100 | 12 | 2 | −3.0 ** | 9 | 2 | −2.1 ** | 8 | 5 | −0.8 ** | 17 | 9 | −0.9 ** | SK | SKP1-like protein |
| MD08G1106600 | 109 | 6 | −4.2 ** | 24 | 15 | −0.7 NS | 58 | 34 | −0.8 ** | 16 | 20 | 0.3 NS | scpl | serine carboxypeptidase |
| MD14G1056600 | 107 | 38 | −1.5 ** | 173 | 139 | −0.3 NS | 132 | 39 | −1.8 ** | 253 | 107 | −1.2 ** | function unknown |
| MD14G1110100 | 82 | 2 | −5.4 ** | 57 | 4 | −3.7 ** | 66 | 26 | −1.3 ** | 82 | 19 | −2.1 ** | function unknown |
| MD15G1297000 | 91 | 10 | −3.3 ** | 80 | 6 | −3.7 ** | 123 | 25 | −2.3 ** | 80 | 24 | −1.7 ** | APK | adenylyl-sulfate kinase |
| MD16G1091200 | 274 | 12 | −4.5 ** | 83 | 7 | −3.5 ** | 291 | 36 | −3.0 ** | 90 | 22 | −2.0 ** | XTH | xyloglucan endotransglucosylase |
| Secondary candidate gene | |||||||||||||
| MD01G1042500 | 4 | 11 | 1.4 NS | 7 | 8 | 0.2 NS | 2 | 6 | 1.4 ** | 4 | 18 | 2.1 ** | ELI | elicitor-activated gene |
| MD02G1057200 | 12 | 16 | 0.4 NS | 17 | 7 | −1.3 * | 8 | 7 | −0.2 NS | 9 | 8 | −0.2 NS | AUX/IAA | auxin-responsive protein |
| MD05G1098700 | 367 | 111 | −1.7 * | 490 | 357 | −0.5 NS | 140 | 138 | 0.0 NS | 288 | 317 | 0.1 NS | LACS | AMP-dependent synthetase |
| MD10G1315100 | 155 | 76 | −1.0 NS | 23 | 9 | −1.3 * | 101 | 120 | 0.2 NS | 16 | 19 | 0.2 NS | XTH | xyloglucan endotransglucosylase |
| MD11G1230200 | 38 | 13 | −1.5 ** | 31 | 18 | −0.7 * | 38 | 32 | −0.3 NS | 71 | 56 | −0.3 NS | function unknown |
| MD15G1203500 | 2823 | 77 | −5.2 * | 179 | 33 | −2.4 NS | 305 | 531 | 0.8 NS | 5 | 15 | 1.6 ** | ACS | ACC synthase |
| MD17G1141200 | 92 | 32 | −1.5 ** | 110 | 43 | −1.4 * | 88 | 75 | −0.2 NS | 80 | 55 | −0.6 NS | GAUT | galacturonosyltransferase |
| (b) Gene ID | At harvest | After storage | At harvest | After storage | Gene function | ||||||||
| HC | MN | Diff. | HC | MN | Diff. | Retain | Lose | Diff. | Retain | Lose | Diff. | ||
| Primary candidate gene | |||||||||||||
| MD00G1036800 | 52 | 123 | 1.2 NS | 24 | 96 | 2.0 * | 44 | 172 | 2.0 ** | 45 | 92 | 1.0 ** | ABCG | ABC transporter G |
| MD01G1213100 | 789 | 1587 | 1.02 NS | 2193 | 7274 | 1.7 ** | 859 | 1041 | 0.3 NS | 2259 | 11492 | 2.3 ** | CHIA | chitinase |
| MD03G1108400 | 62 | 194 | 1.6 ** | 124 | 819 | 2.7 ** | 12 | 37 | 1.6 ** | 134 | 444 | 1.7 ** | GLTP | glycolipid transfer protein |
| MD05G1310400 | 17 | 2357 | 7.1 ** | 23 | 1585 | 6.1 ** | 391 | 2023 | 2.4 ** | 33 | 769 | 4.5 ** | protein E6-like |
| MD05G1313300 | 10 | 5 | −1.0 * | 15 | 80 | 2.4 * | 4 | 8 | 0.9 ** | 11 | 60 | 2.5 ** | function unknown |
| MD06G1233800 | 51 | 146 | 1.52 NS | 78 | 153 | 1.0 * | 90 | 155 | 0.8 ** | 89 | 228 | 1.4 ** | monoacylglycerol lipase-like |
| MD08G1127900 | 8 | 34 | 2.02 NS | 15 | 50 | 1.7 ** | 5 | 23 | 2.3 ** | 10 | 65 | 2.8 ** | AFR | F-box protein AFR-like |
| MD10G1179100 | 3074 | 8577 | 1.52 NS | 1602 | 64580 | 5.3 ** | 308 | 7092 | 4.5 ** | 42086 | 76687 | 0.9 ** | PG | polygalacturonase |
| MD11G1189000 | 460 | 734 | 0.72 NS | 1626 | 3551 | 1.1 * | 272 | 922 | 1.8 ** | 662 | 10298 | 4.0 ** | BG | glucan endo−1,3-β-glucosidase |
| MD12G1164900 | 41 | 208 | 2.32 NS | 22 | 161 | 2.9 ** | 58 | 185 | 1.7 ** | 30 | 84 | 1.5 ** | PPR | pentatricopeptide repeat protein |
| MD12G1183000 | 46 | 66 | 0.52 NS | 80 | 250 | 1.6 ** | 35 | 45 | 0.3 NS | 77 | 138 | 0.8 ** | LURP-one-related 15-like |
| MD13G1112700 | 16 | 8 | −1.0 NS | 9 | 36 | 2.0 ** | 12 | 26 | 1.1 ** | 17 | 27 | 0.7 * | CYP | cytochrome P450 |
| MD15G1023600 | 896 | 817 | −0.1 NS | 1433 | 13417 | 3.2 * | 89 | 500 | 2.5 ** | 2485 | 7947 | 1.7 ** | JMT | jasmonate O-methyltransferase |
| Secondary candidate gene | |||||||||||||
| MD03G1060100 | 72 | 58 | −0.3 NS | 44 | 122 | 1.5 NS | 26 | 41 | 0.6 * | 27 | 83 | 1.6 ** | LBD | LOB domain-containing protein |
| MD05G1297900 | 11 | 40 | 1.9 NS | 11 | 172 | 4.0 ** | 10 | 39 | 1.9 NS | 147 | 140 | −0.1 NS | EFR | EF-TU receptor |
| MD05G1349800 | 1837 | 1358 | −0.4 NS | 2045 | 1726 | −0.2 NS | 1434 | 2284 | 0.7 ** | 1382 | 2055 | 0.6 ** | WRKY | WRKY transcription factor |
| MD06G1090600 | 14 | 45 | 1.7 NS | 46 | 112 | 1.3 NS | 5 | 10 | 1.1 NS | 76 | 507 | 2.7 ** | ACS | ACC synthase |
| MD16G1158300 | 2231 | 5769 | 1.4 NS | 13467 | 23748 | 0.8 NS | 5066 | 7463 | 0.6 NS | 19752 | 27723 | 0.5 * | α -AF | α -arabinofuranosidase |
| MD16G1277800 | 3 | 13 | 2.1 NS | 5 | 10 | 0.9 NS | 3 | 10 | 1.9 ** | 10 | 17 | 0.8 ** | NRT | nitrate transporter |
| MD17G1256100 | 19 | 56 | 1.5 NS | 34 | 67 | 1.0 NS | 27 | 42 | 0.6 NS | 81 | 286 | 1.8 ** | SFBB | F-box family protein |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, H.-Y.; Tong, C.B.S. Identification of Candidate Genes Involved in Fruit Ripening and Crispness Retention Through Transcriptome Analyses of a ‘Honeycrisp’ Population. Plants 2020, 9, 1335. https://doi.org/10.3390/plants9101335
Chang H-Y, Tong CBS. Identification of Candidate Genes Involved in Fruit Ripening and Crispness Retention Through Transcriptome Analyses of a ‘Honeycrisp’ Population. Plants. 2020; 9(10):1335. https://doi.org/10.3390/plants9101335
Chicago/Turabian StyleChang, Hsueh-Yuan, and Cindy B. S. Tong. 2020. "Identification of Candidate Genes Involved in Fruit Ripening and Crispness Retention Through Transcriptome Analyses of a ‘Honeycrisp’ Population" Plants 9, no. 10: 1335. https://doi.org/10.3390/plants9101335
APA StyleChang, H. -Y., & Tong, C. B. S. (2020). Identification of Candidate Genes Involved in Fruit Ripening and Crispness Retention Through Transcriptome Analyses of a ‘Honeycrisp’ Population. Plants, 9(10), 1335. https://doi.org/10.3390/plants9101335