DNA-Based Authentication and Metabolomics Analysis of Medicinal Plants Samples by DNA Barcoding and Ultra-High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry (UHPLC-MS)
Abstract
:1. Introduction
2. Results and Discussion
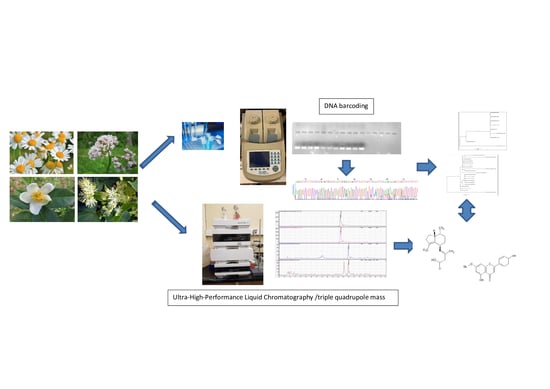
2.1. DNA Barcoding Analysis
2.2. UHPLC/MS Analysis
3. Materials and Methods
3.1. Reagents
3.2. Herbal Products
3.3. DNA Barcoding Analysis
3.3.1. DNA Extraction
3.3.2. PCR and Sequencing
3.3.3. Data Analysis
3.4. UHPLC-MS/MS Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO (World Health Organization). The World Traditional Medicines Situation. In Traditional Medicines: Global Situation, Issues and Challenges; WHO (World Health Organization): Geneva, Switzerland, 2011; pp. 1–14. [Google Scholar]
- De Luca, V.; Salim, V.; Atsumi, S.M.; Yu, F. Mining the biodiversity of plants: A revolution in the making. Science 2012, 336, 1658–1661. [Google Scholar] [CrossRef]
- Debbie, S.; Ladds, G.; Duez, P.; Williamson, E.; Chan, K. Pharmacovigilance of herbal medicine. J. Ethnopharmacol. 2012, 140, 513–518. [Google Scholar]
- Ekor, M. The growing use of herbal medicines: Issues relating to adverse reactions and challenges in monitoring safety. Front. Pharmacol. 2014, 4, 177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Güney, O.I. Consumption attributes and preferences on medicinal and aromatic plants: A consumer segmentation analysis. Cienc. Rural 2019, 49, 5. [Google Scholar] [CrossRef]
- Sofowora, A.; Ogunbodede, E.; Onayade, A. The role and place of medicinal plants in the strategies for disease prevention. Afr. J. Tradit. Complement. Altern. Med. 2013, 10, 210–229. [Google Scholar] [CrossRef] [PubMed]
- Allkin, B. Useful Plants–Medicines: At Least 28,187 Plant Species are Currently Recorded as Being of Medicinal Use. In State of the World’s Plants; Willis, K.J., Ed.; Royal Botanic Gardens: London, UK, 2017. [Google Scholar]
- Wei, S.; Luo, Z.; Cui, S.; Qiao, J.; Zhang, Z.; Zhang, L.; Fu, J.; Ma, X. Molecular Identification and Targeted Quantitative Analysis of Medicinal Materials from Uncaria Species by DNA Barcoding and LC-MS/MS. Molecules 2019, 24, 175. [Google Scholar] [CrossRef] [Green Version]
- Neves, E.O.; de Sales, P.M.; Silveira, D. Pharmacopeial specifications and analytical data from post-marketing quality sampling and testing programs: A perspective beyond out-of-specification results. J. Pharm. Biomed. Anal. 2020, 178, 112935. [Google Scholar] [CrossRef]
- Palhares, R.M.; Gonçalves, M.; Dos Santos, B.; Pereira, G.; das Graças, M.; Oliveira, G. Medicinal plants recommended by the world health organization: DNA barcode identification associated with chemical analyses guarantees their quality. PLoS ONE 2015, 10, e0127866. [Google Scholar] [CrossRef] [Green Version]
- CBOL Plant Working Group. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef] [Green Version]
- Tungmunnithum, D.; Renouard, S.; Drouet, S.; Blondeau, J.P.; Hano, C. A Critical Cross-Species Comparison of Pollen from Nelumbo nucifera Gaertn. vs. Nymphaea lotus L. for Authentication of Thai Medicinal Herbal Tea. Plants 2020, 9, 921. [Google Scholar]
- Pawar, R.S.; Handy, S.M.; Cheng, R.; Shyong, N.; Grundel, E. Assessment of the Authenticity of Herbal Dietary Supplements: Comparison of Chemical and DNA Barcoding Methods. Planta Med. 2017, 83, 921–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abubakar, M.B.; Salleh, M.F.; Shamsir Omar, M.S.; Wagiran, A. DNA Barcoding and Chromatography Fingerprints for the Authentication of Botanicals in Herbal Medicinal Products. Evid. Based Complement Altern. Med. 2017, 1352948. [Google Scholar] [CrossRef]
- Mili Bhargava, M.; Sharma, A. DNA barcoding in plants: Evolution and applications of in silico approaches and resources. Mol. Phylogenet. Evol. 2013, 67, 631–641. [Google Scholar] [CrossRef] [PubMed]
- Ichim, M.C. The DNA-Based Authentication of Commercial Herbal Products Reveals Their Globally Widespread Adulteration. Front. Pharm. 2019, 10, 1227. [Google Scholar] [CrossRef] [PubMed]
- Belščak-Cvitanović, A.; Valinger, D.; Benković, M.; Tušek, A.J.; Jurina, T.; Komes, D.; Kljusurić, J.G. Integrated approach for bioactive quality evaluation of medicinal plant extracts using HPLC-DAD, spectrophotometric, near infrared spectroscopy and chemometric techniques. Int. J. Food Prop. 2017, 20, 2463–2480. [Google Scholar] [CrossRef]
- Zuo, Y.; Chen, Z.; Kondo, K.; Funamoto, T.; Wen, J.; Zhou, S. DNA barcoding of Panax species. Planta Med. 2011, 77, 182–187. [Google Scholar] [CrossRef]
- Jianping, H.; Xiaohui Pang, X.; Baosheng, L.; Hui, Y.; Jingyuan, S.; Shilin, C. An authenticity survey of herbal medicines from markets in China using DNA barcoding. Sci. Rep. 2016, 6, 18723. [Google Scholar]
- Sánchez, M.; González-Burgos, E.; Iglesias, I.; Lozano, R.; Gómez-Serranillos, M.P. Current uses and knowledge of medicinal plants in the Autonomous Community of Madrid (Spain): A descriptive cross-sectional study. BMC Complement. Med. Ther. 2020, 20, 306. [Google Scholar] [CrossRef]
- Newmaster, S.G.; Grguric, M.; Shanmughanandhan, D.; Ramalingam, S.; Ragupathy, S. DNA barcoding detects contamination and substitution in North American herbal products. BMC Med. 2013, 11, 222. [Google Scholar] [CrossRef] [Green Version]
- Wachtel-Galor, S.; Benzie, I.F.F. Herbal Medicine: An Introduction to Its History, Usage, Regulation, Current Trends, and Research Needs. In Herbal Medicine: Biomolecular and Clinical Aspects, 2nd ed.; Benzie, I.F.F., Wachtel-Galor, S., Eds.; CRC Press/Taylor & Francis: Boca Raton, FL, USA, 2011; Chapter 1. [Google Scholar]
- Howard, C.; Lockie-Williams, C.; Slater, A. Applied Barcoding: The Practicalities of DNA Testing for Herbals. Plants 2020, 9, 1150. [Google Scholar] [CrossRef]
- Upton, R.; David, B.; Gafner, S.; Glasl, S. Botanical ingredient identification and quality assessment: Strengths and limitations of analytical techniques. Phytochem. Rev. 2019, 19, 1157–1177. [Google Scholar] [CrossRef] [Green Version]
- Hausmann, A.; Haszprunar, G.; Hebert, P.D.N. DNA Barcoding the Geometrid Fauna of Bavaria (Lepidoptera): Successes, Surprises, and Questions. PLoS ONE 2011, 6, e17134. [Google Scholar] [CrossRef] [PubMed]
- Janjua, S.; Fakhar-i-Abbas; William, K. DNA Mini-barcoding for wildlife trade control: A case study on identification of highly processed animal materials. Mitochondrial DNA 2016, 27, 544–546. [Google Scholar] [CrossRef] [PubMed]
- Ford, C.S.; Ayres, K.L.; Haider, N.; Toomey, N.; van-Alpen-Stahl, J.; Kelly, L.J.; Wikström, N.; Hollingsworth, P.M.; Duff, R.J.; Hoot, S.B.; et al. Selection of candidate DNA barcoding regions for use on land plants. Bot. J. Linn. Soc. 2009, 159, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Raclariu, A.C.; Paltinean, R.; Vlase, L.; Labarre, A.; Manzanilla, V.; Ichim, M.C.; Crisan, G.; Brysting, A.K.; Boer, H. Comparative authentication of Hypericum perforatum herbal products using DNA metabarcoding, TLC and HPLC-MS. Sci. Rep. 2017, 7, 1291. [Google Scholar] [CrossRef] [PubMed]
- Raclariu, A.C.; Ţebrencu, C.E.; Ichim, M.C.; Ciupercǎ, O.T.; Brysting, A.K.; Boer, H. What’s in the box? Authentication of Echinacea herbal products using DNA metabarcoding and HPTLC. Phytomedicine 2018, 15, 32–38. [Google Scholar] [CrossRef]
- Hernández, J.; Raggone, M.; Bonazzola, P.; Bandoni, A.; Consolini, A. Antitussive, antispasmodic, bronchodilating and cardiac inotropic effects of the essential oil from Blepharocalyx salicifolius leaves. J. Ethnopharmacol. 2018, 210, 107–117. [Google Scholar] [CrossRef] [Green Version]
- Ivanova, N.V.; Kuzmina, M.L.; Braukmann, T.W.A.; Borisenko, A.V.; Zakharov, E.V. Authentication of Herbal Supplements Using Next- Generation Sequencing. PLoS ONE 2016, 11, e0156426. [Google Scholar] [CrossRef]
- Techen, N.; Parveen, I.; Pan, Z.; Khan, I.A. DNA barcoding of medicinal plant material for identification. Curr. Opin. Biotechnol. 2014, 25, 103–110. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.; Ye, L.; Kang, Z.; Jia, D.; Yang, L.; Zhang, B. LC-MS-Based Metabolomic Approach Revealed the Significantly Different Metabolic Profiles of Five Commercial Truffle Species. Front. Microbiol. 2019, 10, 2227. [Google Scholar] [CrossRef]
- Saleem, H.; Htar, T.T.; Naidu, R.; Anwar, S.; Zengin, G.; Locatelli, M.; Ahemad, N. HPLC-PDA Polyphenolic Quantification, UHPLC-MS Secondary Metabolite Composition, and In Vitro Enzyme Inhibition Potential of Bougainvillea glabra. Plants 2020, 9, 388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Medicines Agency (EMA). Available online: https://www.ema.europa.eu/en/documents/herbal-report/final-assessment-report-matricaria-recutita-l-flos-matricaria-recutita-l-aetheroleum_en.pdf (accessed on 30 September 2020).
- Real Farmacopea Española. Available online: https://extranet.boe.es/farmacopea/ (accessed on 1 October 2020).
- Haghi, G.; Hatami, A.; Safaei, A.; Mehran, M. Analysis of phenolic compounds in Matricaria chamomilla and its extracts by UPLC-UV. RPS 2014, 9, 31–37. [Google Scholar] [PubMed]
- European Medicines Agency (EMA). Available online: https://www.ema.europa.eu/en/documents/herbal-report/final-assessment-report-valeriana-officinalis-l-radix-valeriana-officinalis-l-aetheroleum_en.pdf (accessed on 30 September 2020).
- Navarrete, A.; Avula, B.; Choi, Y.W.; Khan, I.A. Chemical fingerprinting of valeriana species: Simultaneous determination of valerenic acids, flavonoids, and phenylpropanoids using liquid chromatography with ultraviolet detection. J. AOAC Int. 2006, 89, 8–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Medicines Agency (EMA). Available online: https://www.ema.europa.eu/en/documents/herbal-report/draft-assessment-report-camellia-sinensis-l-kuntze-non-fermentatum-folium_en.pdf (accessed on 1 October 2020).
- European Medicines Agency. Available online: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-specifications-test-procedures-acceptance-criteria-herbal-substances-herbal-preparations/traditional-herbal-medicinal-products-revision-1_en.pdf (accessed on 25 September 2020).
- Raghavendra, H.L.; Kumar, S.V.P.; Kekuda, T.R.P.; Ejeta, E.; Molla, B.; Anilakumar, K.R.; Khanum, F. HPLC method for chemical composition and in vitro antioxidant activity of Camellia sinensis Linn. Anal. Chem. Lett. 2011, 1, 361–369. [Google Scholar] [CrossRef]
- Pieczykolan, A.; Pietrzak, W.; Nowak, R.; Pielczyk, J.; Lamacz, K. Optimization of Extraction Conditions for Determination of Tiliroside in Tilia L. Flowers Using an LC-ESI-MS/MS Method. J. Anal. Methods Chem. 2019, 9052425. [Google Scholar] [CrossRef] [Green Version]
- Nowak, R. Separation and quantification of tiliroside from plant extracts by SPE/RP-HPLC. Pharm. Biol. 2003, 41, 627–630. [Google Scholar] [CrossRef]
- Sayers, E.W.; Barrett, T.; Benson, D.A.; Bolton, E.; Bryant, S.H.; Canese, K.; Chetvernin, V.; Church, D.M.; Dicuccio, M.; Federhen, S.; et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011, 39, D38–D51. [Google Scholar] [CrossRef] [Green Version]
- Pedales, R.D.; Damatac, A.M.; Limbo, C.A.; Marquez, C.M.; Navarro, A.I.B.; Molina, J. DNA Barcoding of Philippine Herbal Medicinal Products. J. AOAC Int. 2016, 99, 1479–1489. [Google Scholar] [CrossRef]
- Katoh, K.; Toh, H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008, 9, 286–298. [Google Scholar] [CrossRef] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A. “FigTree 1.2.2.”. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 13 August 2020).
- Ratnasingham, S. Hebert PDN. BOLD: The Barcode of Life Data System. Mol. Ecol. Notes 2007, 7, 355–364. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Zheng, Z.; Deng, X.; Ma, X.; Wang, S.; Liu, J.; Liu, Y.; Shi, J. Flexible and powerful strategy for qualitative and quantitative analysis of valepotriates in Valeriana jatamansi Jones using high-performance liquid chromatography with linear ion trap Orbitrap mass spectrometry. J. Sep. Sci. 2017, 40, 1906–1919. [Google Scholar] [CrossRef] [PubMed]
- Scoparo, C.T.; de Souza, L.M.; Dartora, N.; Sassaki, G.L.; Gorin, P.A.; Iacomini, M. Analysis of Camellia sinensis green and black teas via ultra high performance liquid chromatography assisted by liquid-liquid partition and two-dimensional liquid chromatography (size exclusion × reversed phase). J. Chromatogr. A 2012, 1222, 29–37. [Google Scholar] [CrossRef] [PubMed]



| Sample Code | Labelled | BOLD System Identification |
|---|---|---|
| MH1 | Matricaria | Matricaria recutita |
| MH2 | Chamomile | Matricaria recutita |
| MH3 | Matricaria chamomilla | Matricaria recutita |
| MF2 | Chamomile | Matricaria recutita |
| MS1 | Chamomile | Matricaria recutita |
| MS2 | Matricaria recutita | Matricaria recutita |
| MS3 | Chamomile | Matricaria recutita |
| CH1 | Camellia sinensis | Blepharocalyx tweediei |
| CH2 | Thea | Camellia sinensis |
| CF3 | Thea | Camellia sinensis |
| VH1 | Valeriana officinalis | Valeriana hirtella |
| VS1 | Valeriana officinalis | Valeriana hirtella |
| VF2 | Valeriana officinalis | Valeriana hirtella |
| TH1 | Tilia platyphyllos | Tilia cordata |
| TH2 | Linden | Tilia cordata |
| TH3 | Tilia Europea | Tilia cordata |
| TS2 | Linden | Malvales |
| Matricaria recutita L. | |
|---|---|
| Sample | Apigenin-7-glucoside (mg/g) (mean ± SD) |
| MH1 | 0.35 ± 0.015 |
| MH2 | 0.02 ± 0.004 |
| MH3 | 0.05 ± 0.005 |
| MS1 | 0.07 ± 0.004 |
| MS2 | 0.06 ± 0.003 |
| MS3 | 0.16 ± 0.009 |
| MF1 | 0.03 ± 0.002 |
| MF2 | 0.01 ± 0.004 |
| MF3 | 0.03 ± 0.005 |
| Valeriana Officinalis L. | ||
|---|---|---|
| Sample | Acetoxyvalerenic Acid (mg/g) (mean ± SD) | Valerenic Acid (mg/g) (mean ± SD) |
| VH1 | 0.26 ± 0.030 | 1.09 ± 0.017 |
| VH2 | 0.24 ± 0.011 | 1.15 ± 0.017 |
| VH3 | 0.24 ± 0.014 | 1.01 ± 0.020 |
| VF1 | 0.20 ± 0.020 | 0.48 ± 0.015 |
| VF2 | 0.29 ± 0.017 | 0.78 ± 0.026 |
| VF3 | 0.53 ± 0.014 | 1.67 ± 0.030 |
| VS1 | 0.25 ± 0.055 | 0.84 ± 0.015 |
| Camellia Sinensis (L.) Kuntze | |
|---|---|
| Sample | Epigallocatechin (mg/g) (mean ± SD) |
| CH1 | 21.2 ± 0.025 |
| CH2 | 47.2 ± 0.015 |
| CH3 | 23.2 ± 0.060 |
| CS1 | 32.9 ± 0.025 |
| CS2 | 37.1 ± 0.011 |
| CF1 | 15.1 ± 0.020 |
| CF2 | 25.9 ± 0.011 |
| CF3 | 25.5 ± 0.030 |
| Tilia spp. | |
|---|---|
| Sample | Tiliroside (mg/g) (mean ± SD) |
| TH1 | 0.32 ± 0.056 |
| TH2 | 0.39 ± 0.045 |
| TH3 | 0.08 ± 0.020 |
| TS1 | 0.27 ± 0.026 |
| TS2 | 0.42 ± 0.032 |
| TS3 | 0.43 ± 0.025 |
| TF1 | 0.23 ± 0.020 |
| TF2 | 0.36 ± 0.010 |
| TF3 | 0.35 ± 0.011 |
| Chamomile | Valerian | ||||
|---|---|---|---|---|---|
| Pharmacies | Herbal Shops | Supermarkets | Pharmacies | Herbal Shops | Supermarkets |
| MF1 | MH1 | MS1 | VF1 | VH1 | VS1 |
| MF2 | MH2 | MS2 | VF2 | VH2 | |
| MF3 | MH3 | MS3 | VF3 | VH3 | |
| Linden | Tea | ||||
| Pharmacies | Herbal Shops | Supermarkets | Pharmacies | Herbal Shops | Supermarkets |
| TF1 | TH1 | TS1 | CF1 | CH1 | CS1 |
| TF2 | TH2 | TS2 | CF2 | CH2 | CS2 |
| TF3 | TH3 | TS3 | CF3 | CH3 | |
| Active Compound | Transition | CE | Dwell (msec) |
|---|---|---|---|
| Acetoxyalerenic acid | 291.3 > 59.00 | 27 V | 100 |
| 291.3 > 249.05 | 17 V | 100 | |
| Valerenic acid | 233.2 > 40.92 | 22 V | 100 |
| 233.2 > 84.20 | 25 V | 100 | |
| Epigallocatechin | 457 > 169.05 | 18 V | 100 |
| 457 > 125.00 | 45 V | 100 | |
| Tiliroside | 592.8 > 284.95 | 33 V | 100 |
| 592.8 > 254.95 | 55V | 100 | |
| Apigenin-7-O-glucoside | 431.3 > 267.90 | 35 V | 100 |
| 431.3 > 150.95 | 52 V | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sánchez, M.; González-Burgos, E.; Divakar, P.K.; Gómez-Serranillos, M.P. DNA-Based Authentication and Metabolomics Analysis of Medicinal Plants Samples by DNA Barcoding and Ultra-High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry (UHPLC-MS). Plants 2020, 9, 1601. https://doi.org/10.3390/plants9111601
Sánchez M, González-Burgos E, Divakar PK, Gómez-Serranillos MP. DNA-Based Authentication and Metabolomics Analysis of Medicinal Plants Samples by DNA Barcoding and Ultra-High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry (UHPLC-MS). Plants. 2020; 9(11):1601. https://doi.org/10.3390/plants9111601
Chicago/Turabian StyleSánchez, Marta, Elena González-Burgos, Pradeep Kumar Divakar, and M. Pilar Gómez-Serranillos. 2020. "DNA-Based Authentication and Metabolomics Analysis of Medicinal Plants Samples by DNA Barcoding and Ultra-High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry (UHPLC-MS)" Plants 9, no. 11: 1601. https://doi.org/10.3390/plants9111601
APA StyleSánchez, M., González-Burgos, E., Divakar, P. K., & Gómez-Serranillos, M. P. (2020). DNA-Based Authentication and Metabolomics Analysis of Medicinal Plants Samples by DNA Barcoding and Ultra-High-Performance Liquid Chromatography/Triple Quadrupole Mass Spectrometry (UHPLC-MS). Plants, 9(11), 1601. https://doi.org/10.3390/plants9111601