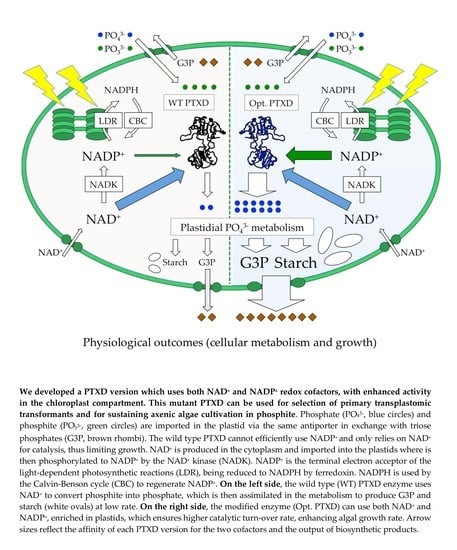
A Phosphite Dehydrogenase Variant with Promiscuous Access to Nicotinamide Cofactor Pools Sustains Fast Phosphite-Dependent Growth of Transplastomic Chlamydomonas reinhardtii
Abstract
:1. Introduction
2. Results
2.1. Site-Directed Mutagenesis of PTXD Cofactor-Binding Amino Acidic Residues
2.2. The Mutagenized PTXD Version Enables Faster Selective Growth of Transplastomic Lines
2.3. Faster Selective Growth is Related to a Higher Catalytic Efficiency of the Mutagenized PTXD Version
2.4. The Optimized PTXD Version Enables Fast and Reliable Recovery of Transformants
3. Discussion
4. Materials and Methods
4.1. Algal Strains and Cultivation Strategies
4.2. Transformation of the Chloroplast Genome of C. reinhardtii
4.3. Transformation of the Nuclear Genome of C. reinhardtii
4.4. Production of Recombinant PTXD Protein in E. coli
4.5. Phenotypical, Genetic and Biochemical Characterization of All Transgenic Lines
4.6. Bioinformatics Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Rasala, B.A.; Mayfield, S.P. Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth. Res. 2014, 123, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Esland, L.; Larrea-Alvarez, M.; Purton, S. Selectable Markers and Reporter Genes for Engineering the Chloroplast of Chlamydomonas reinhardtii. Boilogy 2018, 7, 46. [Google Scholar] [CrossRef] [Green Version]
- Maul, J.E.; Lilly, J.W.; Cui, L.; Depamphilis, C.W.; Miller, W.; Harris, E.H.; Stern, D.B. The Chlamydomonas reinhardtii plastid chromosome: Islands of genes in a sea of repeats. Plant Cell 2002, 14, 2659–2679. [Google Scholar] [CrossRef]
- Doron, L.; Segal, N.; Shapira, M. Transgene Expression in Microalgae—From Tools to Applications. Front. Plant Sci. 2016, 7, 234. [Google Scholar] [CrossRef] [PubMed]
- Verma, D.; Daniell, H. Chloroplast Vector Systems for Biotechnology Applications1. Plant Physiol. 2007, 145, 1129–1143. [Google Scholar] [CrossRef] [Green Version]
- Schroda, M. Good News for Nuclear Transgene Expression in Chlamydomonas. Cells 2019, 8, 1534. [Google Scholar] [CrossRef] [Green Version]
- Almaraz-Delgado, A.L.; Flores-Uribe, J.; Pérez-España, V.H.; Salgado-Manjarrez, E.; Badillo-Corona, J. Production of therapeutic proteins in the chloroplast of Chlamydomonas reinhardtii. AMB Express 2014, 4, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmad, N.; Mehmood, M.A.; Malik, S. Recombinant Protein Production in Microalgae: Emerging Trends. Protein Pept. Lett. 2020, 27, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Carney, L.T.; Lane, T.W. Parasites in algae mass culture. Front. Microbiol. 2014, 5, 278. [Google Scholar] [CrossRef] [Green Version]
- Day, J.G.; Gong, Y.; Hu, Q. Microzooplanktonic grazers—A potentially devastating threat to the commercial success of microalgal mass culture. Algal Res. 2017, 27, 356–365. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, W.; Chen, L.; Wang, J.; Liu, T. The contamination and control of biological pollutants in mass cultivation of microalgae. Bioresour. Technol. 2013, 128, 745–750. [Google Scholar] [CrossRef] [PubMed]
- Lian, J.; Wijffels, R.H.; Smidt, H.; Sipkema, D. The effect of the algal microbiome on industrial production of microalgae. Microb. Biotechnol. 2018, 11, 806–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leão, P.N.; Pereira, A.R.; Liu, W.-T.; Ng, J.; Pevzner, P.A.; Dorrestein, P.C.; König, G.M.; Vasconcelos, V.; Gerwick, W.H. Synergistic allelochemicals from a freshwater cyanobacterium. Proc. Natl. Acad. Sci. USA 2010, 107, 11183–11188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costas, A.M.G.; White, A.K.; Metcalf, W.W. Purification and Characterization of a Novel Phosphorus-oxidizing Enzyme fromPseudomonas stutzeriWM88. J. Boil. Chem. 2001, 276, 17429–17436. [Google Scholar] [CrossRef] [Green Version]
- Loera-Quezada, M.M.; González, M.A.L.; Velázquez-Juárez, G.; Sanchez-Calderón, L.; Nascimento, M.D.; López-Arredondo, D.; Herrera-Estrella, L. A novel genetic engineering platform for the effective management of biological contaminants for the production of microalgae. Plant Biotechnol. J. 2016, 14, 2066–2076. [Google Scholar] [CrossRef] [Green Version]
- Changko, S.; Rajakumar, P.D.; Young, R.E.B.; Purton, S. The phosphite oxidoreductase gene, ptxD as a bio-contained chloroplast marker and crop-protection tool for algal biotechnology using Chlamydomonas. Appl. Microbiol. Biotechnol. 2019, 104, 675–686. [Google Scholar] [CrossRef] [Green Version]
- Sandoval-Vargas, J.M.; Macedo-Osorio, K.S.; Durán-Figueroa, N.V.; Garibay-Orijel, C.; Badillo-Corona, J. Chloroplast engineering of Chlamydomonas reinhardtii to use phosphite as phosphorus source. Algal Res. 2018, 33, 291–297. [Google Scholar] [CrossRef]
- Sandoval-Vargas, J.M.; Jiménez-Clemente, L.A.; Macedo-Osorio, K.S.; Oliver-Salvador, M.C.; Fernández-Linares, L.C.; Durán-Figueroa, N.V.; Badillo-Corona, J. Use of the ptxD gene as a portable selectable marker for chloroplast transformation in Chlamydomonas reinhardtii. Mol. Biotechnol. 2019, 61, 461–468. [Google Scholar] [CrossRef]
- Johannes, T.W.; Woodyer, R.D.; Zhao, H. Directed Evolution of a Thermostable Phosphite Dehydrogenase for NAD(P)H Regeneration. Appl. Environ. Microbiol. 2005, 71, 5728–5734. [Google Scholar] [CrossRef] [Green Version]
- Woodyer, R.; Van Der Donk, W.A.; Zhao, H. Relaxing the Nicotinamide Cofactor Specificity of Phosphite Dehydrogenase by Rational Design†. Biochemistry 2003, 42, 11604–11614. [Google Scholar] [CrossRef]
- Michelet, L.; Burr, S.; Rochaix, J.-D.; Lefebvre-Legendre, L.; Goldschmidt-Clermont, M. Enhanced chloroplast transgene expression in a nuclear mutant of Chlamydomonas. Plant Biotechnol. J. 2010, 9, 565–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldschmidt-Clermont, M. Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of chlamydomonas. Nucleic Acids Res. 1991, 19, 4083–4089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boynton, J.; Gillham, N.; Harris, E.; Hosler, J.; Johnson, A.; Jones, A.; Randolph-Anderson, B.; Robertson, D.; Klein, T.; Shark, K.; et al. Chloroplast transformation in Chlamydomonas with high velocity microprojectiles. Science 1988, 240, 1534–1538. [Google Scholar] [CrossRef] [PubMed]
- Day, A.; Goldschmidt-Clermont, M. The chloroplast transformation toolbox: Selectable markers and marker removal. Plant Biotechnol. J. 2011, 9, 540–553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beacham, T.A.; Sweet, J.B.; Allen, M.J. Large scale cultivation of genetically modified microalgae: A new era for environmental risk assessment. Algal Res. 2017, 25, 90–100. [Google Scholar] [CrossRef]
- Relyea, H.A.; Van Der Donk, W.A. Mechanism and applications of phosphite dehydrogenase. Bioorganic Chem. 2005, 33, 171–189. [Google Scholar] [CrossRef]
- Bisson, C.; Adams, N.; Stevenson, B.; Brindley, A.A.; Polyviou, D.; Bibby, T.S.; Baker, P.; Hunter, C.N.; Hitchcock, A. The molecular basis of phosphite and hypophosphite recognition by ABC-transporters. Nat. Commun. 2017, 8, 1746. [Google Scholar] [CrossRef]
- Weber, A.P. Solute transporters as connecting elements between cytosol and plastid stroma. Curr. Opin. Plant Boil. 2004, 7, 247–253. [Google Scholar] [CrossRef]
- Hashida, S.-N.; Kawai-Yamada, M. Inter-Organelle NAD Metabolism Underpinning Light Responsive NADP Dynamics in Plants. Front. Plant Sci. 2019, 10, 960. [Google Scholar] [CrossRef] [Green Version]
- Hochmal, A.K.; Schulze, S.; Trompelt, K.; Hippler, M. Calcium-dependent regulation of photosynthesis. Biochim. Biophys. Acta (BBA) Bioenerg. 2015, 1847, 993–1003. [Google Scholar] [CrossRef] [Green Version]
- Forti, G.; Furia, A.; Bombelli, P.; Finazzi, G. In Vivo Changes of the Oxidation-Reduction State of NADP and of the ATP/ADP Cellular Ratio Linked to the Photosynthetic Activity in Chlamydomonas reinhardtii1. Plant Physiol. 2003, 132, 1464–1474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bajhaiya, A.K.; Dean, A.; Zeef, L.A.; Webster, R.E.; Pittman, J.K. PSR1 Is a Global Transcriptional Regulator of Phosphorus Deficiency Responses and Carbon Storage Metabolism in Chlamydomonas reinhardtii. Plant Physiol. 2015, 170, 1216–1234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moseley, J.L.; Chang, C.-W.; Grossman, A.R. Genome-Based Approaches to Understanding Phosphorus Deprivation Responses and PSR1 Control in Chlamydomonas reinhardtii†‡. Eukaryot. Cell 2006, 5, 26–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz, F.A.; Marchesini, N.; Seufferheld, M.; Govindjee; Docampo, R. The Polyphosphate Bodies of Chlamydomonas reinhardtiiPossess a Proton-pumping Pyrophosphatase and Are Similar to Acidocalcisomes. J. Boil. Chem. 2001, 276, 46196–46203. [Google Scholar] [CrossRef] [Green Version]
- Werner, T.P.; Amrhein, N.; Freimoser, F. Inorganic polyphosphate occurs in the cell wall of Chlamydomonas reinhardtii and accumulates during cytokinesis. BMC Plant Boil. 2007, 7, 51. [Google Scholar] [CrossRef] [Green Version]
- Irihimovitch, V.; Yehudai-Resheff, S. Phosphate and sulfur limitation responses in the chloroplast of Chlamydomonas reinhardtii. FEMS Microbiol. Lett. 2008, 283, 1–8. [Google Scholar] [CrossRef]
- Johnson, X.; Alric, J. Central Carbon Metabolism and Electron Transport in Chlamydomonas reinhardtii: Metabolic Constraints for Carbon Partitioning between Oil and Starch. Eukaryot. Cell 2013, 12, 776–793. [Google Scholar] [CrossRef] [Green Version]
- Zou, Y.; Zhang, H.; Brunzelle, J.S.; Johannes, T.W.; Woodyer, R.; Hung, J.E.; Nair, N.U.; Van Der Donk, W.A.; Zhao, H.; Nair, S.K. Crystal Structures of Phosphite Dehydrogenase Provide Insights into Nicotinamide Cofactor Regeneration. Biochemistry 2012, 51, 4263–4270. [Google Scholar] [CrossRef] [Green Version]
- Gallaher, S.; Fitz-Gibbon, S.; Glaesener, A.G.; Pellegrini, M.; Merchant, S.S. Chlamydomonas Genome Resource for Laboratory Strains Reveals a Mosaic of Sequence Variation, Identifies True Strain Histories, and Enables Strain-Specific Studies. Plant Cell 2015, 27, 2335–2352. [Google Scholar] [CrossRef]
- Gorman, D.S.; Levine, R.P. Cytochrome f and plastocyanin: Their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc. Natl. Acad. Sci. USA 1965, 54, 1665–1669. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, Y. Codon usage tabulated from international DNA sequence databases: Status for the year 2000. Nucleic Acids Res. 2000, 28, 292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Puigbo, P.; Guzmán, E.; Romeu, A.; Garcia-Vallve, S. OPTIMIZER: A web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007, 35, W126–W131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guzman-Zapata, D.; Macedo-Osorio, K.S.; Almaraz-Delgado, A.L.; Durán-Figueroa, N.; Badillo-Corona, J. Production of Recombinant Proteins in the Chloroplast of the Green Alga Chlamydomonas reinhardtii. Adv. Struct. Saf. Stud. 2016, 1385, 69–85. [Google Scholar]
- Cao, M.; Fu, Y.; Guo, Y.; Pan, J. Chlamydomonas (Chlorophyceae) colony PCR. Protoplasma 2009, 235, 107–110. [Google Scholar] [CrossRef]
- Bertalan, I.; Munder, M.; Weiß, C.; Kopf, J.; Fischer, D.; Johanningmeier, U. A rapid, modular and marker-free chloroplast expression system for the green alga Chlamydomonas reinhardtii. J. Biotechnol. 2015, 195, 60–66. [Google Scholar] [CrossRef]







© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cutolo, E.; Tosoni, M.; Barera, S.; Herrera-Estrella, L.; Dall’Osto, L.; Bassi, R. A Phosphite Dehydrogenase Variant with Promiscuous Access to Nicotinamide Cofactor Pools Sustains Fast Phosphite-Dependent Growth of Transplastomic Chlamydomonas reinhardtii. Plants 2020, 9, 473. https://doi.org/10.3390/plants9040473
Cutolo E, Tosoni M, Barera S, Herrera-Estrella L, Dall’Osto L, Bassi R. A Phosphite Dehydrogenase Variant with Promiscuous Access to Nicotinamide Cofactor Pools Sustains Fast Phosphite-Dependent Growth of Transplastomic Chlamydomonas reinhardtii. Plants. 2020; 9(4):473. https://doi.org/10.3390/plants9040473
Chicago/Turabian StyleCutolo, Edoardo, Matteo Tosoni, Simone Barera, Luis Herrera-Estrella, Luca Dall’Osto, and Roberto Bassi. 2020. "A Phosphite Dehydrogenase Variant with Promiscuous Access to Nicotinamide Cofactor Pools Sustains Fast Phosphite-Dependent Growth of Transplastomic Chlamydomonas reinhardtii" Plants 9, no. 4: 473. https://doi.org/10.3390/plants9040473
APA StyleCutolo, E., Tosoni, M., Barera, S., Herrera-Estrella, L., Dall’Osto, L., & Bassi, R. (2020). A Phosphite Dehydrogenase Variant with Promiscuous Access to Nicotinamide Cofactor Pools Sustains Fast Phosphite-Dependent Growth of Transplastomic Chlamydomonas reinhardtii. Plants, 9(4), 473. https://doi.org/10.3390/plants9040473