The Selection of Gamma-Ray Irradiated Higher Yield Rice Mutants by Directed Evolution Method
Abstract
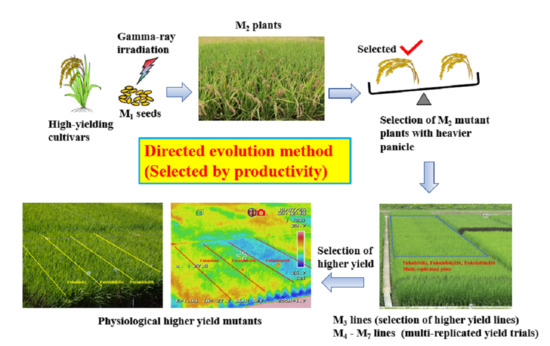
:1. Introduction
2. Results
2.1. Screening of the Candidate Mutant M2 Plants and M3 Lines
2.2. Yield Evaluation of M4 Mutant Lines in 2016
2.3. Yield Evaluation of M5 Mutant Lines in 2017
2.4. Evaluations of Yield and Yield-Related Traits of M5, M6, and M7 Mutant Lines in 2017, 2018, and 2019
2.5. DNA Marker Analysis between Higher Yield Mutants and Their Original Varieties
3. Discussion
3.1. Rice Higher Yield Mutants and Their Gene Identifications
3.2. The Comparison between Higher Yield Mutants Obtained in This Report and Previously Reported High-Yielding Materials
3.3. The Method in This Report Is Considered to Be an Application of the “Directed Evolution Method”
3.4. Physiological Traits as the Underlying Causes behind Higher Yield Mutations
3.5. Some Other Considerations Concerning Our Methods
4. Materials and Methods
4.1. Plant Materials and Irradiation Treatment
4.2. Plant Selection from M2 Populations and Pedigree Selection of M3 to M7 Materials
4.3. Field Evaluations of Higher Yield Mutant Candidates in M4 to M7
4.4. DNA Preparation and SNP Genotyping
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ray, D.K.; Mueller, N.D.; West, P.C.; Foley, J.A. Yield trends are insufficient to double global crop production by 2050. PLoS ONE 2013, 8, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.G. Meeting the global food demand of the future by engineering Crop Photosynthesis and Yield Potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Cao, L.; Sun, X.; Zhu, Y.; Zhang, T.; Jiang, L.; Liu, Y.; Dong, S.; Sun, D.; Yang, J.; et al. Fine mapping of a major quantitative trait locus, qgnp7(t), controlling grain number per panicle in African rice (Oryza glaberrima S.). Breed. Sci. 2018, 68, 606–613. [Google Scholar] [CrossRef] [Green Version]
- Song, X.J.; Huang, W.; Shi, M.; Zhu, M.Z.; Lin, H.X. A QTL for rice grain width and weight encodes a previously unknown RING-type E3. Nat. Genet. 2007, 39, 623–630. [Google Scholar] [CrossRef]
- Shomura, A.; Izawa, T.; Ebana, K.; Ebitani, T.; Kanegae, H.; Konishi, S.; Yano, M. Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 2008, 40, 1023–1028. [Google Scholar] [CrossRef]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.J.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef]
- Ujiie, K.; Yamamoto, T.; Yano, M.; Ishimaru, K. Genetic factors determining varietal differences in characters affecting yield between two rice (Oryza sativa L.) varieties, Koshihikari and IR64. Genet. Resour. Crop Evol. 2016, 63, 97–123. [Google Scholar] [CrossRef]
- Ishimaru, K.; Hirotsu, N.; Madoka, Y.; Murakami, N.; Hara, N.; Onodera, H.; Kashiwagi, T.; Ujiie, K.; Shimizu, B.; Onishi, A.; et al. Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 2013, 45, 707–711. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, M.; Zhang, Q.; Xu, Z.; Xu, Q. The DENSE AND ERECT PANICLE 1 (DEP1) gene offering the potential in the breeding of high-yielding rice. Breed. Sci. 2016, 66, 659–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taguchi-Shiobara, F.; Kawagoe, Y.; Kato, H.; Onodera, H.; Tagiri, A.; Hara, N.; Miyao, A.; Hirochika, H.; Kitano, H.; Yano, H.; et al. A loss-of-function mutation of rice DENSE PANICLE 1 causes semi-dwarfness and slightly increased number of spikelets. Breed. Sci. 2011, 61, 17–25. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Sun, J.; Xiao, Z.; Cheng, F.; Xu, H.; Tang, L.; Chen, W.; Xu, Z.; Xu, Q. Variations in DENSE AND ERECT PANICLE 1 (DEP1) contribute to the diversity of the panicle trait in high-yielding japonica rice varieties in northern China. Breed. Sci. 2016, 66, 599–605. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Qian, Q.; Liu, Z.; Sun, H.; He, S.; Luo, D.; Xia, G.; Chu5, C.; Li, J.; Fu, X. Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 2009, 41, 494–497. [Google Scholar] [CrossRef]
- Liu, J.; Tao, H.J.; Shi, S.; Ye, W.J.; Qian, Q.; Guo, L.B. Genetics and breeding improvement for panicle type in rice. Chin. J. Rice Sci. 2012, 26, 227–234. [Google Scholar]
- Wang, J.; Nakazaki, T.; Chen, S.; Chen, W.; Saito, H.; Tsukiyama, T.; Okumoto, Y.; Xu, X.; Tanisaka, T. Identification and characterization of the erect-pose panicle gene EP conferring high grain yield in rice (Oryza sativa L.). Theor. Appl. Genet. 2009, 119, 85–91. [Google Scholar] [CrossRef]
- Takai, T.; Adachi, S.; Taguchi-Shiobara, F.; Sanoh-Arai, Y.; Iwasawa, N.; Yoshinaga, S.; Hirose, S.; Taniguchi, Y.; Yamanouchi, U.; Wu, J.; et al. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci. Rep. 2013, 3, 1–11. [Google Scholar] [CrossRef]
- Takai, T.; Ohsumi, A.; Arai, Y.; Iwasawa, N.; Yano, M.; Yamamoto, T.; Yoshinaga, Y.; Kondo, M. QTL Analysis of Leaf Photosynthesis in Rice. JARQ 2013, 47, 227–235. [Google Scholar] [CrossRef] [Green Version]
- Fujita, D.; Trijatmiko, K.R.; Tagle, A.G.; Sapasap, M.V.; Koide, Y.; Sasaki, K.; Tsakirpaloglou, N.; Gannaban, R.B.; Nishimura, T.; Yanagihara, S.; et al. NAL1 allele from a rice landrace greatly increases yield in modern indica cultivars. PNAS 2013, 110, 20341–20346. [Google Scholar] [CrossRef] [Green Version]
- Fujita, D.; Koide, Y.; Kobayashi, N. Genetic dissection of agronomic traits in introgression lines and improvement of an elite indica rice variety. JARQ 2018, 52, 91–103. [Google Scholar] [CrossRef] [Green Version]
- Murai, M.; Nakamura, K.; Saito, M.; Nagayama, A.; Ise, K. Yield-increasing effect of a major gene, Ur-1 (Undulated rachis-1) on different genetic background in rice. Breed. Sci. 2005, 55, 279–285. [Google Scholar] [CrossRef] [Green Version]
- Li, H.W.; Hu, C.H.; Chang, W.T.; Weng, T.S. Progress report on the studies of utilization of X-Ray radiation for rice improvement, 1961. Int. J. Appl. Radiat. Isot. 1962, 13, 455–465. [Google Scholar] [CrossRef]
- Li, F.; Shimizu, A.; Nishio, T.; Tsutsumi, N.; Kato, H. Comparison and characterization of mutations induced by gamma-ray and carbon-ion irradiation in rice (Oryza sativa L.) using whole-genome resequencing. G3 2019, 9, 3743–3751. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Numa, H.; Hara, N.; Sentoku, N.; Ishii, T.; Fukuta, Y.; Nishimura, N.; Kato, H. Identification of a locus for seed shattering in rice (Oryza sativa L.) by combining bulked segregant analysis with whole-genome sequencing. Mol. Breed. 2019, 39, 36. [Google Scholar] [CrossRef]
- Fukushima, A.; Ohta, H.; Yokogami, N.; Tsuda, N. Effects of heavy nitrogen fertilization and direct seeding on the yield and grain quality of the rice varieties bred at the tohoku agricultural research center. Jpn. J. Crop Sci. 2017, 86, 339–346. [Google Scholar] [CrossRef]
- Li, F.; Shimizu, S.; Kato, H. Identification of candidate causal genes for higher yield rice mutant lines using a bulk sequencing method. Breed. Res. 2020, 22, 119. [Google Scholar]
- Fujita, D.; Santos, R.E.; Ebron, L.A.; Telebanco-Yanoria, M.J.; Kato, H.; Kobayashi, S.; Uga, Y.; Araki, E.; Takai, T.; Tsunematsu, H.; et al. Development of introgression lines of an Indica-type rice variety, IR64, for unique agronomic traits and detection of the responsible chromosomal regions. Field Crops Res. 2009, 114, 244–254. [Google Scholar] [CrossRef]
- Nakagawa, H.; Kato, H. Induced mutations for food and energy security: Challenge of inducing unique mutants for new cultivars and molecular research. Bull. NARO Crop Sci. 2017, 1, 33–124. [Google Scholar]
- Cobb, R.E.; Chao, R.; Zhao, H. Directed evolution: Past, present and future. AIChE J. 2013, 59, 1432–1440. [Google Scholar] [CrossRef] [Green Version]
- Cobb, R.E.; Sun, N.; Zhao, H. Directed evolution as a powerful synthetic biology tool. Methods 2013, 60, 81–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gionfriddo, M.; Gara, L.D.; Loreto, F. Directed evolution of plant processes: Towards a green (r)evolution? Trends Plant Sci. 2019, 24, 999–1007. [Google Scholar] [CrossRef] [PubMed]
- Butt, H.; Ayman Eid, A.; Momin, A.A.; Bazin, J.; Crespi, M.; Arold, S.T.; Mahfouz, M.M. CRISPR directed evolution of the spliceosome for resistance to splicing inhibitors. Genome Biol. 2019, 20, 73. [Google Scholar] [CrossRef] [Green Version]
- Engqvist, M.K.M.; Rabeb, K.S. Applications of protein engineering and directed evolution in plant research. Plant Physiol. 2019, 179, 907–917. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Name of the Lines or Varieties | Gen. | Heading Date | Maturity Date | Culm Length (cm) | Panicle Length (cm) | No. Panicles (/m2) | Lodging | Air-Dried | Ratio (%) | Brown Rice w. (t/ha) | Ratio (%) | Residual Rice (%) | Seed Set (%) | 1000 b.r.w. (g) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole w. (t/ha) | Grain w. (t/ha) | ||||||||||||||
| FukuhibikiH6 | M4 | Jul.27 | Sept.5 | 64.2 | 18.0 | 306 | 0.0 | 16.0 | 7.07 | 102.6 | 5.92 | 102.6 | 0.2 | 89.4 | 24.0 |
| FukuhibikiH7 | M4 | Jul.26 | Sept.1 | 67.5 | 18.6 | 296 | 0.0 | 15.8 | 6.73 | 97.7 | 5.68 | 98.4 | 0.1 | 88.8 | 23.9 |
| Fukuhibiki | Jul.27 | Sept.1 | 64.7 | 18.7 | 300 | 0.0 | 16.4 | 6.89 | 100.0 | 5.77 | 100.0 | 0.2 | 86.6 | 23.4 | |
| OonariH1 | M4 | Aug.9 | Sept.30 | 72.4 | 27.1 | 268 | 0.5 | 17.1 | 9.62 | 99.3 | 8.15 | 101.4 | 0.1 | 81.9 | 22.2 |
| OonariH2 | M4 | Aug.8 | Sept.28 | 70.5 | 27.1 | 268 | 0.5 | 16.8 | 9.82 | 101.3 | 8.15 | 101.4 | 0.1 | 85.4 | 21.1 |
| Oonari | Aug.8 | Sept.30 | 74.0 | 26.2 | 287 | 0.5 | 17.1 | 9.69 | 100.0 | 8.04 | 100.0 | 0.1 | 80.9 | 21.7 | |
| YamadawaraH1 | M4 | Aug.9 | Sept.30 | 78.0 | 20.6 | 288 | 0.0 | 17.4 | 8.43 | 101.4 | 7.18 | 102.0 | 0.2 | 86.7 | 22.7 |
| YamadawaraH2 | M4 | Aug.9 | Sept.30 | 81.1 | 21.1 | 282 | 0.5 | 17.6 | 8.71 | 104.8 | 7.38 | 104.8 | 0.1 | 85.8 | 22.7 |
| YamadawaraH3 | M4 | Aug.8 | Sept.30 | 81.5 | 21.2 | 297 | 1.0 | 18.2 | 8.93 | 107.5 | 7.52 | 106.8 | 0.2 | 86.5 | 23.4 |
| YamadawaraH4 | M4 | Aug.10 | Sept.30 | 77.5 | 20.9 | 286 | 0.0 | 17.4 | 8.62 | 103.7 | 7.29 | 103.6 | 0.2 | 85.5 | 22.7 |
| Yamadawara | Aug.9 | Sept.30 | 78.3 | 20.8 | 293 | 0.0 | 16.8 | 8.31 | 100.0 | 7.04 | 100.0 | 0.2 | 85.2 | 22.8 | |
| MochidawaraH1 | M4 | Aug.12 | Oct.6 | 84.5 | 26.5 | 208 | 1.0 | 18.1 | 9.91 | 104.4 | 8.16 | 103.3 | 0.1 | 87.7 | 22.6 |
| MochidawaraH2 | M4 | Aug.13 | Oct.6 | 86.9 | 26.4 | 196 | 1.0 | 17.8 | 9.51 | 100.2 | 7.89 | 99.9 | 0.1 | 84.0 | 22.3 |
| Mochidawara | Aug.12 | Oct.6 | 84.5 | 26.2 | 209 | 1.0 | 17.7 | 9.49 | 100.0 | 7.90 | 100.0 | 0.2 | 87.2 | 22.4 | |
| AkidawaraH1 | M4 | Aug.12 | Sept.27 | 77.2 | 20.9 | 291 | 0.5 | 17.7 | 7.96 | 97.9 | 6.78 | 98.1 | 0.1 | 92.4 | 21.0 |
| AkidawaraH2 | M4 | Aug.12 | Sept.27 | 80.7 | 22.3 | 290 | 0.5 | 17.8 | 7.80 | 95.9 | 6.68 | 96.7 | 0.1 | 90.8 | 21.4 |
| Akidawara | Aug.13 | Sept.27 | 76.5 | 21.1 | 291 | 0.5 | 18.6 | 8.13 | 100.0 | 6.91 | 100.0 | 0.1 | 89.3 | 21.7 | |
| Mutant (upper) Original v. (lower) | Gen. | Heading Date | Maturity Date | Culm Length (cm) | Panicle Length (cm) | No. Panicles (/m2) | Lodging | Air-Dried | Ratio (%) | Seed Set (%) | No. Grains/Panicle | 1000 Grain w. (g) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole w. (t/ha) | Grain w. (t/ha) | ||||||||||||
| FukuhibikiH6 | M5 | 24-Jul * | 9-Sep | 70.5 | 21.8 ** | 247 | 0.0 | 13.77 | 7.98 * | 105.1 | 94.2 | 106.2 | 28.8 * |
| Fukuhibiki | 25-Jul | 9-Sep | 70.2 | 21.3 | 246 | 0.0 | 13.40 | 7.59 | 100.0 | 93.4 | 105.5 | 28.1 | |
| YamadawaraH3 | M5 | 16-Aug | 1-Oct | 84.0 * | 20.3 * | 289 | 0.0 | 19.42 | 9.71 ** | 103.7 | 90.5 | 133.9 | 27.6 |
| Yamadawara | 16-Aug | 1-Oct | 82.4 | 20.0 | 289 | 0.0 | 19.64 | 9.36 | 100.0 | 91.2 | 131.8 | 27.2 | |
| MochidawaraH1 | M5 | 23-Aug | 18-Oct | 91.8 | 26.1 | 228 | 0.0 | 29.51 | 10.40 ** | 104.8 | 82.5 ** | 172.4 | 28.3 |
| Mochidawara | 23-Aug | 18-Oct | 92.0 | 26.0 | 221 | 0.0 | 29.18 | 9.92 | 100.0 | 80.1 | 167.2 | 28.4 | |
| Mutant (Upper) Original v. (Lower) | Gen. | Heading Date | Maturity Date | Culm Length (cm) | Panicle Length (cm) | No. Panicles (/m2) | Lodging | Air-Dried | Ratio (%) | Seed Set (%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole w. (t/ha) | Grain w. (t/ha) | ||||||||||
| FukuhibikiH6 | M6 | 28-Jul | 8-Sep | 84.6 ** | 20.4 | 346 | 0.9 | 18.28 | 9.89 * | 101.2 | 91.3 |
| FukuhibikiH8 | M6 | 28-Jul | 8-Sep | 87.0 ** | 21.0 | 354 | 1.8 | 18.64 ** | 10.18 ** | 104.1 | 92.1 |
| Fukuhibiki | 28-Jul | 8-Sep | 82.6 | 20.5 | 365 | 0.9 | 18.19 | 9.77 | 100.0 | 92.4 | |
| YamadawaraH3 | M6 | 10-Aug ** | 2-Oct | 83.3 | 21.4 ** | 343 | 0.0 | 21.53 | 10.18 ** | 102.7 | 84.9 |
| Yamadawara | 13-Aug | 2-Oct | 81.3 | 21.0 | 322 | 0.0 | 21.25 | 9.92 | 100.0 | 84.5 | |
| MochidawaraH1 | M6 | 15-Aug | 6-Oct | 91.4 | 26.5 | 254 ** | 2.6 | 24.81 | 11.01 | 100.5 | 80.8 |
| Mochidawara | 15-Aug | 6-Oct | 91.5 | 26.2 | 266 | 2.4 | 25.19 | 10.95 | 100.0 | 78.2 | |
| Mutant (Upper) Original v. (Lower) | Gen. | Heading Date | Maturity Date | Culm Length (cm) | Panicle Length (cm) | No. Panicles (/m2) | Lodging | Air-Dried | Ratio (%) | Seed Set (%) | No. Grains/Panicle | 1000 Grain w. (g) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Whole w. (t/ha) | Grain w. (t/ha) | ||||||||||||
| (Standard fertilizer applicaton with 100 kg nitrogen/ha) | |||||||||||||
| FukuhibikiH6 | M7 | 29-Jul * | 5-Sep | 70.0 * | 19.4 * | 318 | 1.0 ** | 14.42 | 6.25 ** | 108.1 | 89.1 | 71.9 | 27.4 * |
| Fukuhibiki | 28-Jul | 5-Sep | 68.3 | 18.5 | 312 | 0.0 | 13.97 | 5.78 | 100.0 | 87.5 | 69.5 | 26.7 | |
| FukuhibikiH8 | M7 | 29-Jul * | 5-Sep | 73.2 ** | 18.6 | 306 | 0.6 * | 14.96 | 7.23 ** | 122.5 | 91.2 * | 82.9 ** | 28.5 ** |
| Fukuhibiki | 28-Jul | 5-Sep | 68.5 | 18.5 | 310 | 0.0 | 14.15 | 5.90 | 100.0 | 87.0 | 71.2 | 26.7 | |
| YamadawaraH3 | M7 | 7-Aug | 20-Sep | 81.8 ** | 20.9 * | 322 | 0.3 * | 16.61 | 8.49 | 103.9 | 90.3 | 91.8 | 28.8 ** |
| Yamadawara | 8-Aug | 20-Sep | 78.6 | 20.3 | 323 | 0.0 | 16.17 | 8.17 | 100.0 | 90.3 | 92.8 | 27.3 | |
| (Heavy fertilizer application with 200 kg nitrogen/ha) | |||||||||||||
| FukuhibikiH6 | M7 | 31-Jul | 5-Sep | 82.3 ** | 19.2 * | 404 | 1.9 ** | 17.70 | 8.65 ** | 109.3 | 93.0 ** | 88.9 ** | 24.0 |
| FukuhibikiH8 | M7 | 31-Jul | 5-Sep | 84.6 ** | 20.1 | 412 | 2.3 ** | 17.70 | 9.17 ** | 115.8 | 94.2 ** | 92.3 ** | 24.3 |
| Fukuhibiki | 30-Jul | 5-Sep | 78.0 | 19.8 | 428 | 1.1 | 17.40 | 7.91 | 100.0 | 85.8 | 76.1 | 24.2 | |
| MochidawaraH1 | M7 | 10-Aug | 2-Oct | 95.3 | 27.3 * | 307 | 8.4 | 22.31 | 10.94 | 101.5 | 91.2 * | 130.1 | 27.3 |
| Mochidawara | 10-Aug | 2-Oct | 94.6 | 28.0 | 294 | 8.6 | 21.73 | 10.78 | 100.0 | 88.7 | 133.3 | 27.6 | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, H.; Li, F.; Shimizu, A. The Selection of Gamma-Ray Irradiated Higher Yield Rice Mutants by Directed Evolution Method. Plants 2020, 9, 1004. https://doi.org/10.3390/plants9081004
Kato H, Li F, Shimizu A. The Selection of Gamma-Ray Irradiated Higher Yield Rice Mutants by Directed Evolution Method. Plants. 2020; 9(8):1004. https://doi.org/10.3390/plants9081004
Chicago/Turabian StyleKato, Hiroshi, Feng Li, and Akemi Shimizu. 2020. "The Selection of Gamma-Ray Irradiated Higher Yield Rice Mutants by Directed Evolution Method" Plants 9, no. 8: 1004. https://doi.org/10.3390/plants9081004
APA StyleKato, H., Li, F., & Shimizu, A. (2020). The Selection of Gamma-Ray Irradiated Higher Yield Rice Mutants by Directed Evolution Method. Plants, 9(8), 1004. https://doi.org/10.3390/plants9081004