Peptide-Conjugated Aggregation-Induced Emission Fluorogenic Probe for Glypican-3 Protein Detection and Hepatocellular Carcinoma Cells Imaging
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Synthesis of NDSA-L5
2.3. Cell Culture
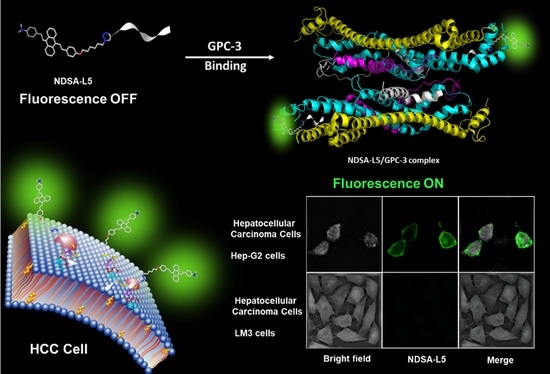
2.4. Cell Imaging
2.5. Cell flow Cytometry Analysis
2.6. In Vivo Imaging
3. Results
3.1. The Photophysical Properties of NDSA-L5
3.2. Cell-Targeted Imaging of NDSA-L5
3.3. In Vivo Imaging in Hep-G2 Cancer Xenograft Mice Model
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Q.Y.; Li, Y.; Min, T.L.; Gong, J.Y.; Du, L.L.; Phillips, D.L.; Liu, J.K.; Lam, J.W.Y.; Sung, H.H.Y.; Williams, I.D.; et al. Time-dependent photodynamic therapy for multiple targets: A highly efficient aie-active photosensitizer for selective bacterial elimination and cancer cell ablation. Angew. Chem. Int. Ed. 2020, 59, 9470–9477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiam-Galvez, K.J.; Allen, B.M.; Spitzer, M.H. Systemic immunity in cancer. Nat. Rev. Cancer 2021, 21, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Mattiuzzi, C.; Lippi, G. Current cancer epidemiology. J. Epidemiol. Glob. Health 2019, 9, 217–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, R.; Mahato, R.I. Recent advances in hepatocellular carcinoma therapy. Pharmacol. Ther. 2017, 173, 106–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weaver, R.J.; Blomme, E.A.; Chadwick, A.E.; Copple, I.M.; Gerets, H.H.J.; Goldring, C.E.; Guillouzo, A.; Hewitt, P.G.; Ingelman-Sundberg, M.; Jensen, K.G.; et al. Managing the challenge of drug-induced liver injury: A roadmap for the development and deployment of preclinical predictive models. Nat. Rev. Drug Discov. 2020, 19, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Vogel, A.; Saborowski, A. Current strategies for the treatment of intermediate and advanced hepatocellular carcinoma. Cancer Treat. Rev. 2020, 82, 101946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anwanwan, D.; Singh, S.K.; Singh, S.; Saikam, V.; Singh, R. Challenges in liver cancer and possible treatment approaches. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188314. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Nault, J.C.; Roberts, L.R.; Zucman-Rossi, J. Genomic medicine and implications for hepatocellular carcinoma prevention and therapy. Gastroenterology 2019, 156, 492–509. [Google Scholar] [CrossRef]
- Rebouissou, S.; Nault, J.C. Advances in molecular classification and precision oncology in hepatocellular carcinoma. J. Hepatol. 2020, 72, 215–229. [Google Scholar] [CrossRef] [Green Version]
- Dimri, M.; Satyanarayana, A. Molecular signaling pathways and therapeutic targets in hepatocellular carcinoma. Cancers 2020, 12, 491. [Google Scholar] [CrossRef] [Green Version]
- Chalasani, N.P.; Porter, K.; Bhattacharya, A.; Book, A.J.; Neis, B.M.; Xiong, K.M.; Ramasubramanian, T.S.; Edwards, V.D.K.; Chen, I.; Johnson, S.; et al. Validation of a novel multitarget blood test shows high sensitivity to detect early stage hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 2022, 20, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Fan, J.L.; Yang, W.; Shen, B.B.; Yang, Y.P.; Zhou, Q.; Chen, W.M.; Daniyal, M.; Xiao, F.; Sheng, W.B.; et al. Endogenous cys-assisted gsh@agncs-rgo nanoprobe for real-time monitoring of dynamic change in gsh levels regulated by natural drug. Anal. Chem. 2020, 92, 1988–1996. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, J.; Sun, L.H.; Zeng, Z.; Zeng, C.; Zeng, F.; Wu, S.Z. Nanoaggregate probe for breast cancer metastasis through multispectral optoacoustic tomography and aggregation-induced nir-i/ii fluorescence imaging. Angew. Chem. Int. Ed. 2020, 59, 10111–10121. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Gao, Y.J.; Ye, S.Y.; Ding, J.N.; Wang, A.N.; Li, P.J.; Shi, H.B. A light-up near-infrared probe with aggregation-induced emission characteristics for highly sensitive detection of alkaline phosphatase. Analyst 2019, 144, 6262–6269. [Google Scholar] [CrossRef]
- Yi, B.; Wu, T.; Zhu, N.; Huang, Y.; Yang, X.Y.; Yuan, L.; Wu, Y.J.; Liang, X.F.; Jiang, X.Q. The clinical significance of ctc enrichment by gpc3-iml and its genetic analysis in hepatocellular carcinoma. J. Nanobiotechnol. 2021, 19, 74. [Google Scholar] [CrossRef]
- Wang, J.Y.; Wang, X.K.; Zhu, G.Z.; Zhou, X.; Yao, J.; Ma, X.P.; Wang, B.; Peng, T. Distinct diagnostic and prognostic values of glypicans gene expression in patients with hepatocellular carcinoma. BMC Cancer 2021, 21, 462. [Google Scholar] [CrossRef]
- Qiang, Z.Y.; Zhang, H.F.; Jin, S.; Yan, C.; Li, Z.; Tao, L.Y.; Yu, H.B. The prognostic value of arginase-1 and glypican-3 expression levels in patients after surgical intrahepatic cholangiocarcinoma resection. World J. Surg. Oncol. 2021, 19, 316. [Google Scholar] [CrossRef]
- Koksal, A.R.; Aydin, Y.; Lin, D.; Nunez, K.; Thevenot, P.; Parsi, M.A.; Cohen, A.J.; Dash, S. Glypican-3 enriched in exosomes not in the microvesicles: A potential biomarker for hepatocellular carcinoma. Hepatology 2021, 74, 691A–692A. [Google Scholar]
- Caviglia, G.P.; Armandi, A.; Rosso, C.; Gaia, S.; Aneli, S.; Rolle, E.; Abate, M.L.; Olivero, A.; Nicolosi, A.; Guariglia, M.; et al. Biomarkers of oncogenesis, adipose tissue dysfunction and systemic inflammation for the detection of hepatocellular carcinoma in patients with nonalcoholic fatty liver disease. Cancers 2021, 13, 2305. [Google Scholar] [CrossRef]
- Au, K.Y.; Chan, K.K.S.; Lo, R.C.L. A clinicopathological study of young-onset hepatocellular carcinoma. Anticancer Res. 2021, 41, 2933–2944. [Google Scholar] [CrossRef]
- Wang, W.Y.; Wei, C. Advances in the early diagnosis of hepatocellular carcinoma. Genes Dis. 2020, 7, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Yao, M.; Yang, J.L.; Wang, L.; Yao, D.F. Carcinoembryonic type specific markers and liver cancer immunotherapy. Zhonghua Gan Zang Bing Za Zhi = Zhonghua Ganzangbing Zazhi = Chin. J. Hepatol. 2020, 28, 466–470. [Google Scholar]
- Li, Z.H.; Cheng, J.; Huang, P.; Song, W.H.; Nong, L.; Huang, L.; Lin, W.Y. Imaging and detection of hepatocellular carcinoma with a hepatocyte-specific fluorescent probe. Anal. Chem. 2022, 94, 3386–3393. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Zheng, Q.C.; Xu, S.; Yuan, Y.; Cheng, X.; Jiang, S.; Kenry; Yu, Q.; Song, Z.; Liu, B.; et al. Theranostic nanodots with aggregation-induced emission characteristic for targeted and image-guided photodynamic therapy of hepatocellular carcinoma. Theranostics 2019, 9, 1264–1279. [Google Scholar] [CrossRef]
- Natarajan, A.; Zhang, H.; Ye, W.; Huttad, L.; Tan, M.D.; Chua, M.S.; Gambhir, S.S.; So, S.K. A humanized anti-gpc3 antibody for immuno-positron emission tomography imaging of orthotopic mouse model of patient-derived hepatocellular carcinoma xenografts. Cancers 2021, 13, 3977. [Google Scholar] [CrossRef]
- Kelada, O.J.; Gutsche, N.T.; Bell, M.; Berman, R.M.; Baidoo, K.E.; Warner, B.M.; Szajek, L.P.; Hong, J.; Ho, M.; Choyke, P.; et al. Immunopet as stoichiometric sensor for glypican-3 in models of hepatocellular carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2020, 108, S180–S181. [Google Scholar] [CrossRef]
- Xu, R.; Wang, J.; Huang, X.Y.; Zhang, Q.Y.; Xie, Y.J.; Pang, L.; Bai, L.C.; Zhou, J.L. Clinical value of spectral ct imaging combined with afp in identifying liver cancer and hepatic focal nodular hyperplasia. J. Buon 2019, 24, 1429–1434. [Google Scholar]
- Chikhaliwala, P.; Chandra, S. Poly-amidoamine dendrimers@Fe3O4 based electrochemiluminescent nanomaterials for biosensing of liver cancer biomarkers. Electroanalysis 2020, 32, 2404–2414. [Google Scholar] [CrossRef]
- Tian, R.; Zhu, L.; Qin, Z.N.; Wang, G.H.; Wang, J.J.; Zhang, H. Glypican-3 (gpc3) targeted fe3o4 core/au shell nanocomplex for fluorescence/mri/photoacoustic imaging-guided tumor photothermal therapy. Biomater. Sci. 2019, 7, 5258–5269. [Google Scholar] [CrossRef]
- Park, J.O.; Stephen, Z.; Sun, C.; Veiseh, O.; Kievit, F.M.; Fang, C.; Leung, M.; Mok, H.; Zhang, M.Q. Glypican-3 targeting of liver cancer cells using multifunctional nanoparticles. Mol. Imaging 2011, 10, 69–77. [Google Scholar] [CrossRef]
- Zhu, D.L.; Qin, Y.S.; Wang, J.J.; Zhang, L.W.; Zou, S.J.; Zhu, X.H.; Zhu, L. Novel glypican-3-binding peptide for in vivo hepatocellular carcinoma fluorescent imaging. Bioconjug. Chem. 2016, 27, 831–839. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Ma, K.; Xu, B.; Tian, W.J. Tunable supramolecular interactions of aggregation-induced emission probe and graphene oxide with biomolecules: An approach toward ultrasensitive label-free and “turn-on” DNA sensing. Small 2016, 12, 6613–6622. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Zhang, F.L.; Sayyadi, N.; Chen, W.J.; Anwer, A.G.; Care, A.; Xu, B.; Tian, W.J.; Goldys, E.M.; Liu, G.Z. “Turn-on” fluorescent aptasensor based on aiegen labeling for the localization of ifn-gamma in live cells. ACS Sens. 2018, 3, 320–326. [Google Scholar] [CrossRef] [PubMed]
- La Lee, Y.; Ahn, B.C.; Lee, Y.; Lee, S.W.; Cho, J.Y.; Lee, J. Targeting of hepatocellular carcinoma with glypican-3-targeting peptide ligand. J. Pept. Sci. 2011, 17, 763–769. [Google Scholar] [CrossRef]
- Jing, J.B.; Xue, Y.R.; Liu, Y.X.; Xu, B.; Li, H.W.; Liu, L.J.; Wu, Y.Q.; Tian, W.J. Co-assembly of hpv capsid proteins and aggregation-induced emission fluorogens for improved cell imaging. Nanoscale 2020, 12, 5501–5506. [Google Scholar] [CrossRef]
- Han, W.K.; Zhang, S.; Qian, J.Y.; Zhang, J.X.; Wang, X.H.; Xie, Z.G.; Xu, B.; Han, Y.Q.; Tian, W.J. Redox-responsive fluorescent nanoparticles based on diselenide-containing aiegens for cell imaging and selective cancer therapy. Chem. Asian J. 2019, 14, 1745–1753. [Google Scholar] [CrossRef]
- Zhang, S.; Ma, L.; Ma, K.; Xu, B.; Liu, L.J.; Tian, W.J. Label-free aptamer-based biosensor for specific detection of chloramphenicol using aie probe and graphene oxide. ACS Omega 2018, 3, 12886–12892. [Google Scholar] [CrossRef]
- Han, H.-H.; Qiu, Y.-J.; Shi, Y.-Y.; Wen, W.; He, X.-P.; Dong, L.-W.; Tan, Y.-X.; Long, Y.-T.; Tian, H.; Wang, H.-Y. Glypican-3-targeted precision diagnosis of hepatocellular carcinoma on clinical sections with a supramolecular 2d imaging probe. Theranostics 2018, 8, 3268–3274. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Jing, J.; Meng, L.; Xu, B.; Ma, X.; Tian, W. Peptide-Conjugated Aggregation-Induced Emission Fluorogenic Probe for Glypican-3 Protein Detection and Hepatocellular Carcinoma Cells Imaging. Chemosensors 2022, 10, 195. https://doi.org/10.3390/chemosensors10050195
Zhang S, Jing J, Meng L, Xu B, Ma X, Tian W. Peptide-Conjugated Aggregation-Induced Emission Fluorogenic Probe for Glypican-3 Protein Detection and Hepatocellular Carcinoma Cells Imaging. Chemosensors. 2022; 10(5):195. https://doi.org/10.3390/chemosensors10050195
Chicago/Turabian StyleZhang, Song, Jiangbo Jing, Lingchen Meng, Bin Xu, Xibo Ma, and Wenjing Tian. 2022. "Peptide-Conjugated Aggregation-Induced Emission Fluorogenic Probe for Glypican-3 Protein Detection and Hepatocellular Carcinoma Cells Imaging" Chemosensors 10, no. 5: 195. https://doi.org/10.3390/chemosensors10050195
APA StyleZhang, S., Jing, J., Meng, L., Xu, B., Ma, X., & Tian, W. (2022). Peptide-Conjugated Aggregation-Induced Emission Fluorogenic Probe for Glypican-3 Protein Detection and Hepatocellular Carcinoma Cells Imaging. Chemosensors, 10(5), 195. https://doi.org/10.3390/chemosensors10050195