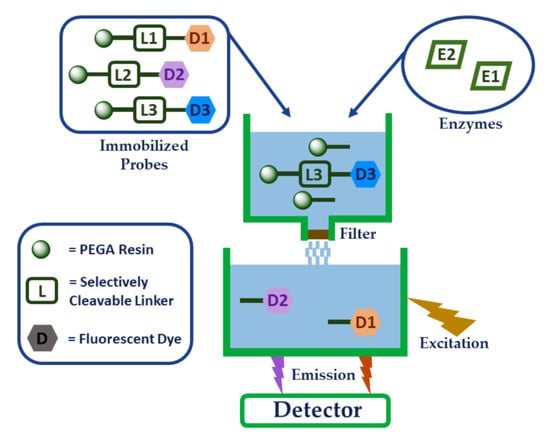
Fluorescence-Based On-Resin Detection of Three Model Proteases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Synthesis of Thrombin Probe on Amino PEGA Resin
2.2. Loading and Yield Determination
2.3. Isolation and Characterization of Thrombin Probe
2.4. Proteases and Inhibitor
2.5. In-Solution Protease Assays
2.6. On-Resin Protease Assays
2.7. Stability Testing of Amino PEGA Resin-Anchored Thrombin Probe
3. Results and Discussion
3.1. Synthesis of Thrombin Probe
3.2. In-Solution Enzymatic Cleavage
3.3. Fluorescence Spectral Properties of Thrombin Probe
3.4. On-Resin Enzyme Studies
3.4.1. The Lowest Detectable Concentration of Thrombin
3.4.2. Simultaneous Detection of Trypsin and Chymotrypsin in the Presence of Thrombin
3.4.3. Detection of Thrombin in the Presence of Trypsin and Chymotrypsin
3.4.4. Stability Testing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ong, I.L.H.; Yang, K.L. Recent developments in protease activity assays and sensors. Analyst 2017, 142, 1867–1881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okorochenkova, Y.; Porubský, M.; Benická, S.; Hlaváč, J. A novel three-fluorophore system as a ratiometric sensor for multiple protease detection. Chem. Commun. 2018, 54, 7589–7592. [Google Scholar] [CrossRef]
- Xu, J.; Fang, L.; Shi, M.; Huang, Y.; Yao, L.; Zhao, S.; Zhang, L.; Liang, H. A peptide-based four-color fluorescent polydopamine nanoprobe for multiplexed sensing and imaging of proteases in living cells. Chem. Commun. 2019, 55, 1651–1654. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, Y.; Guan, X. Simultaneous detection of multiple proteases using a non-array nanopore platform. Nanoscale 2021, 13, 13658–13664. [Google Scholar] [CrossRef] [PubMed]
- Kominami, K.; Nagai, T.; Sawasaki, T.; Tsujimura, Y.; Yashima, K.; Sunaga, Y.; Tsuchimochi, M.; Nishimura, J.; Chiba, K.; Nakabayashi, J.; et al. In Vivo Imaging of Hierarchical Spatiotemporal Activation of Caspase-8 during Apoptosis. PLoS ONE 2012, 7, e50218. [Google Scholar] [CrossRef]
- Li, S.Y.; Liu, L.H.; Cheng, H.; Li, B.; Qiu, W.X.; Zhang, X.Z. A dual-FRET-based fluorescence probe for the sequential detection of MMP-2 and caspase-3. Chem. Commun. 2015, 51, 14520–14523. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, X.; Yuan, S.; Wang, L.; Guan, X. Joint Entropy-Assisted Graphene Oxide-Based Multiplexing Biosensing Platform for Simultaneous Detection of Multiple Proteases. Anal. Chem. 2020, 92, 15042–15049. [Google Scholar] [CrossRef]
- Bui, H.; Brown, C.W.; Buckhout-White, S.; Díaz, S.A.; Stewart, M.H.; Susumu, K.; Oh, E.; Ancona, M.G.; Goldman, E.R.; Medintz, I.L. Transducing Protease Activity into DNA Output for Developing Smart Bionanosensors. Small 2019, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Adem, S.; Jain, S.; Sveiven, M.; Zhou, X.; O’Donoghue, A.J.; Hall, D.A. Giant magnetoresistive biosensors for real-time quantitative detection of protease activity. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Park, S.; Kim, G.; Seo, J.; Yang, H. Ultrasensitive protease sensors using selective affinity binding, selective proteolytic reaction, and proximitY-dependent electrochemical reaction. Anal. Chem. 2016, 88, 11995–12000. [Google Scholar] [CrossRef]
- Chen, H.; Gal, Y.S.; Kim, S.H.; Choi, H.J.; Oh, M.C.; Lee, J.; Koh, K. Potassium ion sensing using a self-assembled calix[4]crown monolayer by surface plasmon resonance. Sens. Actuators B Chem. 2008, 133, 577–581. [Google Scholar] [CrossRef]
- Yang, L.; Wu, T.; Fu, C.; Chen, G.; Xu, S.; Xu, W. SERS determination of protease through a particle-on-a-film configuration constructed by electrostatic assembly in an enzymatic hydrolysis reaction. RSC Adv. 2016, 6, 90120–90125. [Google Scholar] [CrossRef]
- Yoon, H.K.; Yoo, T.H. A novel protease activity assay method based on an engineered autoinhibited protein using an enzyme-linked immunoassay. Analyst 2013, 138, 7164–7168. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.H.; Yang, K.L. Oligopeptide immobilization strategy for improving stability and sensitivity of liquid-crystal protease assays. Sens. Actuators B Chem. 2014, 204, 734–740. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, J.; Gao, Y.; Liu, S.; Koh, K.; Zhu, X.; Yin, Y. Sensitive cell apoptosis assay based on caspase-3 activity detection with graphene oxide-assisted electrochemical signal amplification. Biosens. Bioelectron. 2015, 68, 777–782. [Google Scholar] [CrossRef] [PubMed]
- Swisher, L.Z.; Prior, A.M.; Shishido, S.; Nguyen, T.A.; Hua, D.H.; Li, J. Quantitative electrochemical detection of cathepsin B activity in complex tissue lysates using enhanced AC voltammetry at carbon nanofiber nanoelectrode arrays. Biosens. Bioelectron. 2014, 56, 129–136. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Yu, J.; Bo, B.; Shu, Y.; Li, G. A simple and general approach to assay protease activity with electrochemical technique. Biosens. Bioelectron. 2013, 45, 1–5. [Google Scholar] [CrossRef]
- Xia, N.; Peng, P.; Wang, S.; Du, J.; Zhu, G.; Du, W.; Liu, L. A signal-on electrochemical strategy for protease detection based on the formation of ATCUN-Cu(II). Sens. Actuators B Chem. 2016, 232, 557–563. [Google Scholar] [CrossRef]
- Fan, G.-C.; Han, L.; Zhu, H.; Zhang, J.-R.; Zhu, J.-J. Ultrasensitive Photoelectrochemical Immunoassay for Matrix Metalloproteinase-2 Detection Based on CdS:Mn/CdTe Cosensitized TiO2 Nanotubes and Signal Amplification of SiO2@Ab2 Conjugates. Anal. Chem. 2014, 86, 12398–12405. [Google Scholar] [CrossRef]
- Mu, C.J.; LaVan, D.A.; Langer, R.S.; Zetter, B.R. Self-assembled gold nanoparticle molecular probes for detecting proteolytic activity in vivo. ACS Nano 2010, 4, 1511–1520. [Google Scholar] [CrossRef] [Green Version]
- Olson, E.S.; Jiang, T.; Aguilera, T.A.; Nguyen, Q.T.; Ellies, L.G.; Scadeng, M.; Tsien, R.Y. Activatable cell penetrating peptides linked to nanoparticles as dual probes for in vivo fluorescence and MR imaging of proteases. Proc. Natl. Acad. Sci. USA 2010, 107, 4311–4316. [Google Scholar] [CrossRef] [Green Version]
- Liang, R.P.; Tian, X.C.; Qiu, P.; Qiu, J.D. Multiplexed electrochemical detection of trypsin and chymotrypsin based on distinguishable signal nanoprobes. Anal. Chem. 2014, 86, 9256–9263. [Google Scholar] [CrossRef] [PubMed]
- Milićević, D.; Hlaváč, J. Immobilized fluorescent probes for simultaneous multiple protease detection. Chemosensors 2021, 9, 119. [Google Scholar] [CrossRef]
- Gong, Y.J.; Zhang, X.B.; Mao, G.J.; Su, L.; Meng, H.M.; Tan, W.; Feng, S.; Zhang, G. A unique approach toward near-infrared fluorescent probes for bioimaging with remarkably enhanced contrast. Chem. Sci. 2016, 7, 2275–2285. [Google Scholar] [CrossRef] [Green Version]
- He, G.; Guo, D.; He, C.; Zhang, X.; Zhao, X.; Duan, C. A color-tunable europium complex emitting three primary colors and white light. Angew. Chemie-Int. Ed. 2009, 48, 6132–6135. [Google Scholar] [CrossRef]
- Smith, D.B.; Johnson, K.S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene 1988, 67, 31–40. [Google Scholar] [CrossRef]
- Lanchantin, G.F.; Friedmann, J.A.; Hart, D.W. Interaction of soybean trypsin inhibitor with thrombin and its effect on prothrombin activation. J. Biol. Chem. 1969, 244, 865–875. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Milićević, D.; Hlaváč, J. Fluorescence-Based On-Resin Detection of Three Model Proteases. Chemosensors 2021, 9, 359. https://doi.org/10.3390/chemosensors9120359
Milićević D, Hlaváč J. Fluorescence-Based On-Resin Detection of Three Model Proteases. Chemosensors. 2021; 9(12):359. https://doi.org/10.3390/chemosensors9120359
Chicago/Turabian StyleMilićević, David, and Jan Hlaváč. 2021. "Fluorescence-Based On-Resin Detection of Three Model Proteases" Chemosensors 9, no. 12: 359. https://doi.org/10.3390/chemosensors9120359
APA StyleMilićević, D., & Hlaváč, J. (2021). Fluorescence-Based On-Resin Detection of Three Model Proteases. Chemosensors, 9(12), 359. https://doi.org/10.3390/chemosensors9120359