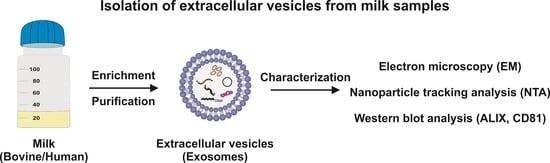
Isolation of Bovine and Human Milk Extracellular Vesicles
Abstract
:1. Introduction
2. Experimental Design
3. Materials and Equipment
3.1. Sources of Milk
3.1.1. Bovine Milk
3.1.2. Human Breast Milk
3.2. Buffers and Solutions
3.2.1. 200 mM HEPES Buffer (pH 7.4)
3.2.2. 50 mM HEPES Buffer (pH 7.4)
3.2.3. 500 mM EDTA Solution (pH 7.5)
3.2.4. Protein Lysis Buffer
3.3. Consumables
3.3.1. Sterile Plastic Tubes with Screw Caps (50 mL)
3.3.2. Sterile Ultracentrifuge Tubes (14 mL)
3.3.3. Plastic Syringes (50 mL with Luer Lock Fitting)
3.3.4. Syringe Filters (with Pore Size 0.20 µm)
3.3.5. Serological Pipets (10 mL and 25 mL)
3.3.6. Lint-Free Cloths
3.3.7. Sterile Scalpels
3.4. Instrumentation
3.4.1. Standard Refrigerated Laboratory Table Centrifuge
3.4.2. Refrigerated Floor Centrifuge with Appropriate Rotor and Bottles
3.4.3. Ultracentrifuge with Appropriate Rotor
3.4.4. Variable Micropipettor and Suitable Sterile Tips
3.5. Antibodies
3.5.1. CD81 Antibody
3.5.2. ALIX Antibody
3.5.3. β-Actin
4. Detailed Procedure
4.1. Removal of Fat Globules, Cream, Cellular Debris, and Somatic Cells
- Transfer the milk into two (100 mL bovine) tubes or one (15 mL human) 50 mL tube.
- Centrifuge the tubes in a standard refrigerated table centrifuge for 30 min at 3000× g. Note: This step allows the removal of fat globules and cream that will float after centrifugation on the supernatant.
- Remove the fat globule/cream fraction that is on the top of the supernatant (Figure 5) with a sterile cube.

- After the removal of the fat globule/cream fraction, transfer the defatted milk solution into suitable centrifuge tubes.
- Centrifuge the tubes in a floor centrifuge at 10,000× g for 30 min. Note: (i) This step allows the removal of cellular debris and somatic cells. (ii) In our laboratory, we transferred the solution to 250 mL polypropylene bottles (2 for bovine samples, and 1 for human sample) and centrifuged the bottles in a Beckman Avanti J-25 high-speed centrifuge (Beckman Coulter, Krefeld, Germany) equipped with a fixed JLA 10.500 angle rotor (10,000× g = 7334 RCF) (Figure 6).

- Transfer the supernatants into sterile 50 mL centrifuge bottles (bovine: 3 tubes; human: 1 tube) and discard the pellets.
- Estimate the volume in each tube.
4.2. Solubilisation of Casein-Containing Protein Aggregates
- Add 500 mM EDTA (pH 7.5) to each tube to reach a final EDTA concentration of 125 mM (i.e., add 3.33 mL per 10 mL defatted milk solution).

- Transfer the solution into 250 mL centrifuge bottles (bovine: 2; human: 1) and centrifuge the tubes in a floor centrifuge at 10,000× g for 30 min as described above.
4.3. Precipitation of Exosomes via Ultracentrifugation
- Transfer the resulting translucid supernatant to sterile ultracentrifuge tubes (bovine: 6; human: 1) and discard the pellet(s). Note: (i) Please read the safety instructions and refer to the instrument instructions before performing any ultracentrifuge run. (ii) In most devices, it is necessary to hook all the buckets, loaded or empty, to the rotor when operating. (iii) Fill the tubes almost to the top edge to prevent the collision of the tubes during the centrifuge run.
- Prepare the tare tubes. Note: (i) In the case of the human sample, prepare a tare tube. (ii) In our laboratory, we use a Beckman OptimaTM L-70K ultracentrifuge (Beckman Coulter, Krefeld, Germany) equipped with an SW 40 Ti rotor (round per minute (rpm) = 29,500; average relative centrifugal force (RCFavg) = 109,895; maximal relative centrifugal force (RCFmax) = 154,779; k-factor: 252.5) to pellet the exosomes. (iii) If you work with another ultracentrifuge or rotor, please refer to the handbook of your instrumentation for g-force to rpm conversion, or alternatively calculate the necessary rpm for the respective ultracentrifuge according to the following equation:
- Centrifuge the tubes at 100,000× g for 70 min.
- Remove the remaining fluid within tubes by placing the liquid-free pellet-containing tubes onto a sterile, absorbent, lint-free cloth paper (Figure 8C).

- Resuspend the clear exosome-enriched pellet carefully into 50 mL (bovine) or 15 mL (human) 200 mM HEPES (pH 7.4). Note: The pellets are gelatinous and not easy to dissolve. Avoid a harsh treatment and minimize the creation of foam during this step. This will damage the exosomes. Be sure that the pellets are well solved before performing the next step.
4.4. Filtration and Washing of Exosomes
- Remove the plunger stopper from the housing of a sterile 50 mL syringe.
- Fix the luer lock of the syringe housing onto a sterile 0.20 µm filter.
- Transfer the exosome-enriched solution into the syringe housing.
- Carefully insert the plunger of the syringe and filter the solution.
- Collect the flow in a sterile 50 mL tube (Figure 9).

- Repeat the filtration step. Use a new sterile 50 mL syringe and a new 0.20 µm filter.
- Transfer the resulting supernatant to 3 (human) sterile ultracentrifuge tubes or 1 (human) tube and repeat the ultracentrifugation step in Section 4.3. After centrifugation, the supernatants should look transparent (Figure 10A).
- Carefully remove the supernatant and remove remaining fluid within tubes by transferring the liquid-free pellet-containing tubes onto a sterile, absorbent, lint-free cloth paper (Figure 10B).

- Cut the bottoms of the tubes with a sterile sharp, thin-bladed scalpel (Figure 11). Note: This step has a high risk of injury. Be careful and make sure that the blade does not break off while cutting. Safety glasses may be worn for protection when cutting.

- Resuspend the pellets of all gradients and combine them in a final volume of 500 µL (bovine) or 100 µL (human) 200 mM HEPES (pH 7.4) buffer. If the exosomes are analyzed via Western blot analysis, resuspend the pellets in protein lysis buffer instead of 200 mM HEPES (pH 7.4).
4.5. Characterisation of Exosome Preparations
4.5.1. Analysis of Exosomal Marker Proteins via Western Blot Analysis
4.5.2. Determination of Size and Concentration of Exosomes via Nanoparticle Tracking Analysis
4.5.3. Imaging of Purified Exosomes via Transmission Electron Microscopy
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Melnik, B.C. Milk—A nutrient system of mammalian evolution promoting mTORC1-dependent translation. Int. J. Mol. Sci. 2015, 16, 17048–17087. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; Stremmel, W.; Weiskirchen, R.; John, S.M.; Schmitz, G. Exosome-derived microRNAs of human milk and their effects on infant health and development. Biomolecules 2021, 11, 851. [Google Scholar] [CrossRef]
- Go, G.; Jeon, J.; Lee, G.; Lee, J.H.; Lee, S.H. Bovine milk extracellular vesicles induce the proliferation and differentiation of osteoblasts and promote osteogenesis in rats. J. Food Biochem. 2021, 45, e13705. [Google Scholar] [CrossRef] [PubMed]
- Melnik, B.C.; John, S.M.; Carrera-Bastos, P.; Schmitz, G. Milk: A postnatal imprinting system stabilizing FoxP3 expression and regulatory T cell differentiation. Clin. Transl. Allergy 2016, 6, 18. [Google Scholar] [CrossRef]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best. Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filén, J.J.; Lahesmaa, R.; Norman, M.; Neve, E.P.; Scheynius, A.; Gabrielsson, S. Exosomes with immune modulatory features are present in human breast milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef]
- Ngu, A.; Wang, S.; Wang, H.; Khanam, A.; Zempleni, J. Milk exosomes in nutrition and drug delivery. Am. J. Physiol. Cell Physiol. 2022, 322, C865–C874. [Google Scholar] [CrossRef]
- Adriano, B.; Cotto, N.M.; Chauhan, N.; Jaggi, M.; Chauhan, S.C.; Yallapu, M.M. Milk exosomes: Nature’s abundant nanoplatform for theranostic applications. Bioact. Mater. 2021, 6, 2479–2490. [Google Scholar] [CrossRef]
- Ohta, M.; Koshida, S.; Jimbo, I.; Oda, M.; Inoue, R.; Tsukahara, T.; Terahara, M.; Nakamura, Y.; Maruo, Y. Highest concentration of breast-milk-derived exosomes in colostrum. Pediatr. Int. 2022, 64, e15346. [Google Scholar] [CrossRef]
- García-Martínez, J.; Pérez-Castillo, Í.M.; Salto, R.; López-Pedrosa, J.M.; Rueda, R.; Girón, M.D. Beneficial effects of bovine milk exosomes in metabolic interorgan cross-talk. Nutrients 2022, 14, 1442. [Google Scholar] [CrossRef]
- Pluchino, S.; Smith, J.A. Explicating exosomes: Reclassifying the rising stars of intercellular communication. Cell 2019, 177, 225–227. [Google Scholar] [CrossRef]
- Ji, G.; Feng, S.; Ren, H.; Chen, W.; Chen, R. Exosomes released from macrophages infected with Talaromyces marneffei activate the innate immune responses and decrease the replication. Immun. Inflamm. Dis. 2023, 11, e881. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, H.; Wang, X.; Lu, M.; Ding, Q.; Chen, A.F.; Xiang, M.; Chen, S. iPSC-derived exosomes promote angiogenesis in naturally aged mice. Aging 2023, 15, 5854–5872. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Teng, Y.; Yang, J.; Yang, L. The role of exosomes in adult neurogenesis: Implications for neurodegenerative diseases. Neural Regen. Res. 2024, 19, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Yasodha, K.; Lizha Mary, L.; Surajit, P.; Satish, R. Exosomes from metastatic colon cancer cells drive the proliferation and migration of primary colon cancer through increased expression of cancer stem cell markers CD133 and DCLK1. Tissue Cell 2023, 84, 102163. [Google Scholar] [CrossRef] [PubMed]
- Tang, N.; Gong, X.R.; Huang, H.; Meng, Q. Activated neutrophil-derived exosomes contribute to blood-brain barrier damage and hemorrhagic transformation after cerebral ischemia/reperfusion. Brain Res. 2023, 1810, 148374. [Google Scholar] [CrossRef] [PubMed]
- Khasabova, I.A.; Khasabov, S.G.; Johns, M.; Juliette, J.; Zheng, A.; Morgan, H.; Flippen, A.; Allen, K.; Golovko, M.Y.; Golovko, S.A.; et al. Exosome-associated lysophosphatidic acid signaling contributes to cancer pain. Pain 2023, 10, 1097. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, M.; Zheng, X.; He, Y.; Ma, X.; Li, J.; Pu, K. Application of exosome engineering modification in targeted delivery of therapeutic drugs. Biochem. Pharmacol. 2023, 215, 115691. [Google Scholar] [CrossRef]
- Fu, P.; Yin, S.; Cheng, H.; Xu, W.; Jiang, J. Engineered exosomes for drug delivery in cancer therapy: A promising approach and application. Curr. Drug Deliv. 2023. ahead of print. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in exosome isolation techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Yamauchi, M.; Shimizu, K.; Rahman, M.; Ishikawa, H.; Takase, H.; Ugawa, S.; Okada, A.; Inoshima, Y. Efficient method for isolation of exosomes from raw bovine milk. Drug Dev. Ind. Pharm. 2019, 45, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Marsh, S.R.; Pridham, K.J.; Jourdan, J.; Gourdie, R.G. Novel protocols for scalable production of high quality purified small extracellular vesicles from bovine milk. Nanotheranostics 2021, 5, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Yamada, T.; Inoshima, Y.; Matsuda, T.; Ishiguro, N. Comparison of methods for isolating exosomes from bovine milk. J. Vet. Med. Sci. 2012, 74, 1523–1525. [Google Scholar] [CrossRef]
- Vaswani, K.; Mitchell, M.D.; Holland, O.J.; Qin Koh, Y.; Hill, R.J.; Harb, T.; Davies, P.S.W.; Peiris, H. A method for the isolation of exosomes from human and bovine milk. J. Nutr. Metab. 2019, 2019, 5764740. [Google Scholar] [CrossRef]
- Wijenayake, S.; Eisha, S.; Tawhidi, Z.; Pitino, M.A.; Steele, M.A.; Fleming, A.S.; McGowan, P.O. Comparison of methods for pre-processing, exosome isolation, and RNA extraction in unpasteurized bovine and human milk. PLoS ONE 2021, 16, e0257633. [Google Scholar] [CrossRef]
- Ko, M.; Kim, H.J.; Park, J.; Lee, H.; Lee, K.N.; Kim, K.; Lee, J.; Yoon, S.J.; Kim, T.; Jeong, S.; et al. Isolation of bovine milk exosome using electrophoretic oscillation assisted tangential flow filtration with antifouling of micro-ultrafiltration membrane filters. ACS Appl. Mater. Interfaces 2023, 15, 26069–26080. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Xie, F.; Cui, D. Exploring purification methods of exosomes from different biological samples. Biomed. Res. Int. 2023, 2023, 2336536. [Google Scholar] [CrossRef]
- Sedykh, S.E.; Purvinsh, L.V.; Burkova, E.E.; Dmitrenok, P.S.; Ryabchikova, E.I.; Nevinsky, G.A. Analysis of proteins and peptides of highly purified CD9+ and CD63+ horse milk exosomes isolated by affinity chromatography. Int. J. Mol. Sci. 2022, 23, 16106. [Google Scholar] [CrossRef]
- Kaddour, H.; Lyu, Y.; Shouman, N.; Mohan, M.; Okeoma, C.M. Development of novel high-resolution size-guided turbidimetry-enabled particle purification liquid chromatography (PPLC): Extracellular vesicles and membraneless condensates in focus. Int. J. Mol. Sci. 2020, 21, 5361. [Google Scholar] [CrossRef]
- Lin, S.H.; Leong, S.L.; Dewan, R.K.; Bloomfield, V.A.; Morr, C.V. Effect of calcium ion on the structure of native bovine casein micelles. Biochemistry 1972, 11, 1818–1821. [Google Scholar] [CrossRef]
- Zigler, J.S., Jr.; Lepe-Zuniga, J.L.; Vistica, B.; Gery, I. Analysis of the cytotoxic effects of light-exposed HEPES-containing culture medium. In Vitro Cell Dev. Biol. 1985, 21, 282–287. [Google Scholar] [CrossRef]
- Vences-Catalán, F.; Rajapaksa, R.; Kuo, C.C.; Miller, C.L.; Lee, A.; Ramani, V.C.; Jeffrey, S.S.; Levy, R.; Levy, S. Targeting the tetraspanin CD81 reduces cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2021, 118, e2018961118. [Google Scholar] [CrossRef]
- Lee, S.S.; Won, J.H.; Lim, G.J.; Han, J.; Lee, J.Y.; Cho, K.O.; Bae, Y.K. A novel population of extracellular vesicles smaller than exosomes promotes cell proliferation. Cell Commun. Signal. 2019, 17, 95. [Google Scholar] [CrossRef]
- Mercier, V.; Laporte, M.H.; Destaing, O.; Blot, B.; Blouin, C.M.; Pernet-Gallay, K.; Chatellard, C.; Saoudi, Y.; Albiges-Rizo, C.; Lamaze, C.; et al. ALG-2 interacting protein-X (Alix) is essential for clathrin-independent endocytosis and signaling. Sci. Rep. 2016, 6, 26986. [Google Scholar] [CrossRef] [PubMed]
- Larios, J.; Mercier, V.; Roux, A.; Gruenberg, J. ALIX- and ESCRT-III-dependent sorting of tetraspanins to exosomes. J. Cell Biol. 2020, 219, e201904113. [Google Scholar] [CrossRef] [PubMed]
- Shu, S.; Yang, Y.; Allen, C.L.; Hurley, E.; Tung, K.H.; Minderman, H.; Wu, Y.; Ernstoff, M.S. Purity and yield of melanoma exosomes are dependent on isolation method. J. Extracell. Vesicles. 2019, 9, 1692401. [Google Scholar] [CrossRef]
- Abramowicz, A.; Marczak, L.; Wojakowska, A.; Zapotoczny, S.; Whiteside, T.L.; Widlak, P.; Pietrowska, M. Harmonization of exosome isolation from culture supernatants for optimized proteomics analysis. PLoS ONE 2018, 13, e0205496. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef]
- Salomon, C.; Ryan, J.; Sobrevia, L.; Kobayashi, M.; Ashman, K.; Mitchell, M.; Rice, G.E. Exosomal signaling during hypoxia mediates microvascular endothelial cell migration and vasculogenesis. PLoS ONE 2013, 8, e68451. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Gupta, A.; Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflam. 2014, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Fenix, A.M.; Franklin, J.L.; Higginbotham, J.N.; Zhang, Q.; Zimmerman, L.J.; Liebler, D.C.; Ping, J.; Liu, Q.; Evans, R.; et al. Reassessment of exosome composition. Cell 2019, 177, 428–445.e18. [Google Scholar] [CrossRef] [PubMed]
- Holliday, L.S.; Faria, L.P.; Rody, W.J., Jr. Actin and actin-associated proteins in extracellular vesicles shed by osteoclasts. Int. J. Mol. Sci. 2019, 21, 158. [Google Scholar] [CrossRef] [PubMed]
- Borkham-Kamphorst, E.; Van de Leur, E.; Meurer, S.K.; Buhl, E.M.; Weiskirchen, R. N-glycosylation of Lipocalin 2 is not required for secretion or exosome targeting. Front. Pharmacol. 2018, 9, 426. [Google Scholar] [CrossRef] [PubMed]
- Kowal, E.J.K.; Ter-Ovanesyan, D.; Regev, A.; Church, G.M. Extracellular vesicle isolation and analysis by Western blotting. Methods Mol. Biol. 2017, 1660, 143–152. [Google Scholar] [CrossRef]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef]
- Schröder, S.K.; Tag, C.G.; Kessel, J.C.; Antonson, P.; Weiskirchen, R. Immunohistochemical detection of estrogen receptor-beta (ERβ) with PPZ0506 antibody in murine tissue: From pitfalls to optimization. Biomedicines 2022, 10, 3100. [Google Scholar] [CrossRef]
- Sokolova, V.; Ludwig, A.K.; Hornung, S.; Rotan, O.; Horn, P.A.; Epple, M.; Giebel, B. Characterisation of exosomes derived from human cells by nanoparticle tracking analysis and scanning electron microscopy. Colloids Surf. B Biointerfaces 2011, 87, 146–150. [Google Scholar] [CrossRef]
- Oosthuyzen, W.; Sime, N.E.; Ivy, J.R.; Turtle, E.J.; Street, J.M.; Pound, J.; Bath, L.E.; Webb, D.J.; Gregory, C.D.; Bailey, M.A.; et al. Quantification of human urinary exosomes by nanoparticle tracking analysis. J. Physiol. 2013, 591, 5833–5842. [Google Scholar] [CrossRef]
- Vestad, B.; Llorente, A.; Neurauter, A.; Phuyal, S.; Kierulf, B.; Kierulf, P.; Skotland, T.; Sandvig, K.; Haug, K.B.F.; Øvstebø, R. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: A variation study. J. Extracell. Vesicles 2017, 6, 1344087. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine 2011, 7, 780–788. [Google Scholar] [CrossRef] [PubMed]
- Samuel, M.; Chisanga, D.; Liem, M.; Keerthikumar, S.; Anand, S.; Ang, C.S.; Adda, C.G.; Versteegen, E.; Jois, M.; Mathivanan, S. Bovine milk-derived exosomes from colostrum are enriched with proteins implicated in immune response and growth. Sci. Rep. 2017, 7, 5933. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Witwer, K.W.; Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef] [PubMed]
- Benmoussa, A.; Gotti, C.; Bourassa, S.; Gilbert, C.; Provost, P. Identification of protein markers for extracellular vesicle (EV) subsets in cow’s milk. J. Proteomics 2019, 192, 78–88. [Google Scholar] [CrossRef]
- Jung, M.K.; Mun, J.Y. Sample preparation and imaging of exosomes by transmission electron microscopy. J. Vis. Exp. 2018, 131, 56482. [Google Scholar] [CrossRef]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weiskirchen, R.; Schröder, S.K.; Weiskirchen, S.; Buhl, E.M.; Melnik, B. Isolation of Bovine and Human Milk Extracellular Vesicles. Biomedicines 2023, 11, 2715. https://doi.org/10.3390/biomedicines11102715
Weiskirchen R, Schröder SK, Weiskirchen S, Buhl EM, Melnik B. Isolation of Bovine and Human Milk Extracellular Vesicles. Biomedicines. 2023; 11(10):2715. https://doi.org/10.3390/biomedicines11102715
Chicago/Turabian StyleWeiskirchen, Ralf, Sarah K. Schröder, Sabine Weiskirchen, Eva Miriam Buhl, and Bodo Melnik. 2023. "Isolation of Bovine and Human Milk Extracellular Vesicles" Biomedicines 11, no. 10: 2715. https://doi.org/10.3390/biomedicines11102715
APA StyleWeiskirchen, R., Schröder, S. K., Weiskirchen, S., Buhl, E. M., & Melnik, B. (2023). Isolation of Bovine and Human Milk Extracellular Vesicles. Biomedicines, 11(10), 2715. https://doi.org/10.3390/biomedicines11102715