The Potential Role of the T2 Ribonucleases in TME-Based Cancer Therapy
Abstract
:1. Introduction
2. Literature Search Strategy
3. T2 RNases: General Features
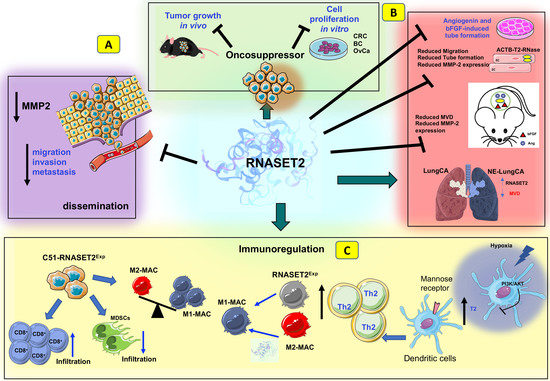
4. T2 RNases: Biological Roles
5. T2 RNases as Evolutionarily Conserved Tumor Suppressors
6. T2 RNases as Modulators of Angiogenesis and Immune Responses
7. RNASET2-Mediated Tumor Suppression by TME Modulation In Vivo
8. Discussion
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Evan, G.I.; Vousden, K.H. Proliferation, cell cycle and apoptosis in cancer. Nature 2001, 411, 342–348. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, T.L. The tumor microenvironment and its role in promoting tumor growth. Oncogene 2008, 27, 5904–5912. [Google Scholar] [CrossRef] [Green Version]
- Balkwill, F.R.; Capasso, M.; Hagemann, T. The tumor microenvironment at a glance. J. Cell Sci. 2012, 125, 5591–5596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baghban, R.; Roshangar, L.; Jahanban-Esfahlan, R.; Seidi, K.; Ebrahimi-Kalan, A.; Jaymand, M.; Kolahian, S.; Javaheri, T.; Zare, P. Tumor microenvironment complexity and therapeutic implications at a glance. Cell Commun. Signal 2020, 18, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ping, Q.; Yan, R.; Cheng, X.; Wang, W.; Zhong, Y.; Hou, Z.; Shi, Y.; Wang, C.; Li, R. Cancer-associated fibroblasts: Overview, progress, challenges, and directions. Cancer Gene Ther. 2021, 28, 984–999. [Google Scholar] [CrossRef]
- Tsoumakidou, M. The advent of immune stimulating CAFs in cancer. Nat. Rev. Cancer 2023, 23, 258–269. [Google Scholar] [CrossRef]
- De Palma, M.; Biziato, D.; Petrova, T.V. Microenvironmental regulation of tumour angiogenesis. Nat. Rev. Cancer 2017, 17, 457–474. [Google Scholar] [CrossRef]
- Carmeliet, P.; Jain, R.K. Angiogenesis in cancer and other diseases. Nature 2000, 407, 249–257. [Google Scholar] [CrossRef]
- Binnewies, M.; Roberts, E.W.; Kersten, K.; Chan, V.; Fearon, D.F.; Merad, M.; Coussens, L.M.; Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Hedrick, C.C.; et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018, 24, 541–550. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnson, D.E.; O’Keefe, R.A.; Grandis, J.R. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 234–248. [Google Scholar] [CrossRef]
- Kitamura, H.; Ohno, Y.; Toyoshima, Y.; Ohtake, J.; Homma, S.; Kawamura, H.; Takahashi, N.; Taketomi, A. Interleukin-6/STAT3 signaling as a promising target to improve the efficacy of cancer immunotherapy. Cancer Sci. 2017, 108, 1947–1952. [Google Scholar] [CrossRef] [Green Version]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFbeta biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 9–34. [Google Scholar] [CrossRef] [PubMed]
- Massague, J. TGFbeta in Cancer. Cell 2008, 134, 215–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allard, B.; Allard, D.; Buisseret, L.; Stagg, J. The adenosine pathway in immuno-oncology. Nat. Rev. Clin. Oncol. 2020, 17, 611–629. [Google Scholar] [CrossRef]
- Stagg, J.; Smyth, M.J. Extracellular adenosine triphosphate and adenosine in cancer. Oncogene 2010, 29, 5346–5358. [Google Scholar] [CrossRef] [Green Version]
- Corbet, C.; Feron, O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 2017, 17, 577–593. [Google Scholar] [CrossRef]
- Certo, M.; Tsai, C.H.; Pucino, V.; Ho, P.C.; Mauro, C. Lactate modulation of immune responses in inflammatory versus tumour microenvironments. Nat. Rev. Immunol. 2021, 21, 151–161. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Tang, T.; Huang, X.; Zhang, G.; Hong, Z.; Bai, X.; Liang, T. Advantages of targeting the tumor immune microenvironment over blocking immune checkpoint in cancer immunotherapy. Signal Transduct. Target. Ther. 2021, 6, 72. [Google Scholar] [CrossRef]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef]
- Jin, M.Z.; Jin, W.L. The updated landscape of tumor microenvironment and drug repurposing. Signal Transduct. Target. Ther. 2020, 5, 166. [Google Scholar] [CrossRef]
- Robert, C. A decade of immune-checkpoint inhibitors in cancer therapy. Nat. Commun. 2020, 11, 3801. [Google Scholar] [CrossRef]
- Darvin, P.; Toor, S.M.; Sasidharan Nair, V.; Elkord, E. Immune checkpoint inhibitors: Recent progress and potential biomarkers. Exp. Mol. Med. 2018, 50, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Jacob, J.B.; Jacob, M.K.; Parajuli, P. Review of immune checkpoint inhibitors in immuno-oncology. Adv. Pharmacol. 2021, 91, 111–139. [Google Scholar] [CrossRef]
- Chen, Y.; McAndrews, K.M.; Kalluri, R. Clinical and therapeutic relevance of cancer-associated fibroblasts. Nat. Rev. Clin. Oncol. 2021, 18, 792–804. [Google Scholar] [CrossRef]
- Saw, P.E.; Chen, J.; Song, E. Targeting CAFs to overcome anticancer therapeutic resistance. Trends Cancer 2022, 8, 527–555. [Google Scholar] [CrossRef]
- Kuyumcu, S.; Sanli, Y.; Subramaniam, R.M. Fibroblast-Activated Protein Inhibitor PET/CT: Cancer Diagnosis and Management. Front. Oncol. 2021, 11, 758958. [Google Scholar] [CrossRef]
- Santana-Viera, L.; Ibba, M.L.; Rotoli, D.; Catuogno, S.; Esposito, C.L. Emerging Therapeutic RNAs for the Targeting of Cancer Associated Fibroblasts. Cancers 2020, 12, 1365. [Google Scholar] [CrossRef]
- Wu, P.; Han, J.; Gong, Y.; Liu, C.; Yu, H.; Xie, N. Nanoparticle-Based Drug Delivery Systems Targeting Tumor Microenvironment for Cancer Immunotherapy Resistance: Current Advances and Applications. Pharmaceutics 2022, 14, 1990. [Google Scholar] [CrossRef]
- Guo, J.; Zeng, H.; Chen, Y. Emerging Nano Drug Delivery Systems Targeting Cancer-Associated Fibroblasts for Improved Antitumor Effect and Tumor Drug Penetration. Mol. Pharm. 2020, 17, 1028–1048. [Google Scholar] [CrossRef]
- Lindner, T.; Giesel, F.L.; Kratochwil, C.; Serfling, S.E. Radioligands Targeting Fibroblast Activation Protein (FAP). Cancers 2021, 13, 5744. [Google Scholar] [CrossRef]
- Shahvali, S.; Rahiman, N.; Jaafari, M.R.; Arabi, L. Targeting fibroblast activation protein (FAP): Advances in CAR-T cell, antibody, and vaccine in cancer immunotherapy. Drug Deliv. Transl. Res. 2023, 13, 2041–2056. [Google Scholar] [CrossRef]
- Xin, L.; Gao, J.; Zheng, Z.; Chen, Y.; Lv, S.; Zhao, Z.; Yu, C.; Yang, X.; Zhang, R. Fibroblast Activation Protein-alpha as a Target in the Bench-to-Bedside Diagnosis and Treatment of Tumors: A Narrative Review. Front. Oncol. 2021, 11, 648187. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Langer, R.; Ferrara, N. Targeting angiogenesis in oncology, ophthalmology and beyond. Nat. Rev. Drug Discov. 2023, 22, 476–495. [Google Scholar] [CrossRef]
- Lopes-Coelho, F.; Martins, F.; Pereira, S.A.; Serpa, J. Anti-Angiogenic Therapy: Current Challenges and Future Perspectives. Int. J. Mol. Sci. 2021, 22, 3765. [Google Scholar] [CrossRef]
- Ansari, M.J.; Bokov, D.; Markov, A.; Jalil, A.T.; Shalaby, M.N.; Suksatan, W.; Chupradit, S.; Al-Ghamdi, H.S.; Shomali, N.; Zamani, A.; et al. Cancer combination therapies by angiogenesis inhibitors; a comprehensive review. Cell Commun. Signal 2022, 20, 49. [Google Scholar] [CrossRef]
- Lee, W.S.; Yang, H.; Chon, H.J.; Kim, C. Combination of anti-angiogenic therapy and immune checkpoint blockade normalizes vascular-immune crosstalk to potentiate cancer immunity. Exp. Mol. Med. 2020, 52, 1475–1485. [Google Scholar] [CrossRef]
- Cerezo, M.; Rocchi, S. Cancer cell metabolic reprogramming: A keystone for the response to immunotherapy. Cell Death Dis. 2020, 11, 964. [Google Scholar] [CrossRef]
- Zhang, D.; Li, J.; Wang, F.; Hu, J.; Wang, S.; Sun, Y. 2-Deoxy-D-glucose targeting of glucose metabolism in cancer cells as a potential therapy. Cancer Lett. 2014, 355, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Hegde, P.S.; Chen, D.S. Top 10 Challenges in Cancer Immunotherapy. Immunity 2020, 52, 17–35. [Google Scholar] [CrossRef] [PubMed]
- Kimiko Sato, F.E. Studies on Ribonucleases in Takadiastase I. J. Biochem. 1957, 44, 753–767. [Google Scholar] [CrossRef]
- Fujio Egami, K.N. Microbial Ribonucleases; Springer: Berlin/Heidelberg, Germany, 1969; Volume 6, pp. 1–92. [Google Scholar] [CrossRef]
- Irie, M. Structure-function relationships of acid ribonucleases: Lysosomal, vacuolar, and periplasmic enzymes. Pharmacol. Ther. 1999, 81, 77–89. [Google Scholar] [CrossRef]
- Deshpande, R.A.; Shankar, V. Ribonucleases from T2 family. Crit. Rev. Microbiol. 2002, 28, 79–122. [Google Scholar] [CrossRef]
- Cannistraro, V.J.; Kennell, D. RNase I*, a form of RNase I, and mRNA degradation in Escherichia coli. J. Bacteriol. 1991, 173, 4653–4659. [Google Scholar] [CrossRef] [Green Version]
- Thompson, D.M.; Parker, R. The RNase Rny1p cleaves tRNAs and promotes cell death during oxidative stress in Saccharomyces cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef]
- McClure, B.A.; Haring, V.; Ebert, P.R.; Anderson, M.A.; Simpson, R.J.; Sakiyama, F.; Clarke, A.E. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature 1989, 342, 955–957. [Google Scholar] [CrossRef]
- Singh, A.; Ai, Y.; Kao, T.H. Characterization of Ribonuclease Activity of Three S-Allele-Associated Proteins of Petunia inflata. Plant Physiol. 1991, 96, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parry, S.; Newbigin, E.; Currie, G.; Bacic, A.; Oxley, D. Identification of active-site histidine residues of a self-incompatibility ribonuclease from a wild tomato. Plant Physiol. 1997, 115, 1421–1429. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Huang, J.; Zheng, Q.; Xie, L.; Lu, X.; Jin, J.; Wang, G. Mammalian mitochondrial RNAs are degraded in the mitochondrial intermembrane space by RNASET2. Protein Cell 2017, 8, 735–749. [Google Scholar] [CrossRef] [Green Version]
- Vidalino, L.; Monti, L.; Haase, A.; Moro, A.; Acquati, F.; Taramelli, R.; Macchi, P. Intracellular trafficking of RNASET2, a novel component of P-bodies. Biol. Cell 2012, 104, 13–21. [Google Scholar] [CrossRef]
- Campomenosi, P.; Salis, S.; Lindqvist, C.; Mariani, D.; Nordstrom, T.; Acquati, F.; Taramelli, R. Characterization of RNASET2, the first human member of the Rh/T2/S family of glycoproteins. Arch. Biochem. Biophys. 2006, 449, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Woodward, J.R.; Bacic, A.; Jahnen, W.; Clarke, A.E. N-Linked Glycan Chains on S-Allele-Associated Glycoproteins from Nicotiana alata. Plant Cell 1989, 1, 511–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shcherbik, N. Golgi-mediated glycosylation determines residency of the T2 RNase Rny1p in Saccharomyces cerevisiae. Traffic 2013, 14, 1209–1227. [Google Scholar] [CrossRef] [PubMed]
- Thorn, A.; Steinfeld, R.; Ziegenbein, M.; Grapp, M.; Hsiao, H.H.; Urlaub, H.; Sheldrick, G.M.; Gartner, J.; Kratzner, R. Structure and activity of the only human RNase T2. Nucleic Acids Res. 2012, 40, 8733–8742. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, H.; Inokuchi, N.; Koyama, T.; Watanabe, H.; Iwama, M.; Ohgi, K.; Irie, M. Primary structure of a base non-specific and adenylic acid preferential ribonuclease from the fruit bodies of Lentinus edodes. Biosci. Biotechnol. Biochem. 1992, 56, 2003–2010. [Google Scholar] [CrossRef]
- Kurihara, H.; Mitsui, Y.; Ohgi, K.; Irie, M.; Mizuno, H.; Nakamura, K.T. Crystal and molecular structure of RNase Rh, a new class of microbial ribonuclease from Rhizopus niveus. FEBS Lett. 1992, 306, 189–192. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, N.; Arai, J.; Inokuchi, N.; Koyama, T.; Ohgi, K.; Irie, M.; Nakamura, K.T. Crystal structure of a plant ribonuclease, RNase LE. J. Mol. Biol. 2000, 298, 859–873. [Google Scholar] [CrossRef]
- Suzuki, A.; Yao, M.; Tanaka, I.; Numata, T.; Kikukawa, S.; Yamasaki, N.; Kimura, M. Crystal structures of the ribonuclease MC1 from bitter gourd seeds, complexed with 2′-UMP or 3′-UMP, reveal structural basis for uridine specificity. Biochem. Biophys. Res. Commun. 2000, 275, 572–576. [Google Scholar] [CrossRef]
- Padmanabhan, S.; Zhou, K.; Chu, C.Y.; Lim, R.W.; Lim, L.W. Overexpression, biophysical characterization, and crystallization of ribonuclease I from Escherichia coli, a broad-specificity enzyme in the RNase T2 family. Arch. Biochem. Biophys. 2001, 390, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Ida, K.; Norioka, S.; Yamamoto, M.; Kumasaka, T.; Yamashita, E.; Newbigin, E.; Clarke, A.E.; Sakiyama, F.; Sato, M. The 1.55 A resolution structure of Nicotiana alata S(F11)-RNase associated with gametophytic self-incompatibility. J. Mol. Biol. 2001, 314, 103–112. [Google Scholar] [CrossRef] [PubMed]
- de Leeuw, M.; Roiz, L.; Smirnoff, P.; Schwartz, B.; Shoseyov, O.; Almog, O. Binding assay and preliminary X-ray crystallographic analysis of ACTIBIND, a protein with anticarcinogenic and antiangiogenic activities. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007, 63, 716–719. [Google Scholar] [CrossRef]
- Luhtala, N.; Parker, R. T2 Family ribonucleases: Ancient enzymes with diverse roles. Trends Biochem. Sci. 2010, 35, 253–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacIntosh, G.C. RNase T2 Family: Enzymatic Properties, Functional Diversity, and Evolution of Ancient Ribonucleases. In Ribonucleases; Springer: Berlin/Heidelberg, Germany, 2011; pp. 89–114. [Google Scholar] [CrossRef]
- Luu, D.T.; Qin, X.; Morse, D.; Cappadocia, M. S-RNase uptake by compatible pollen tubes in gametophytic self-incompatibility. Nature 2000, 407, 649–651. [Google Scholar] [CrossRef]
- Baranzini, N.; Pedrini, E.; Girardello, R.; Tettamanti, G.; de Eguileor, M.; Taramelli, R.; Acquati, F.; Grimaldi, A. Human recombinant RNASET2-induced inflammatory response and connective tissue remodeling in the medicinal leech. Cell Tissue Res. 2017, 368, 337–351. [Google Scholar] [CrossRef]
- Galiana, E.; Bonnet, P.; Conrod, S.; Keller, H.; Panabieres, F.; Ponchet, M.; Poupet, A.; Ricci, P. RNase activity prevents the growth of a fungal pathogen in tobacco leaves and increases upon induction of systemic acquired resistance with elicitin. Plant Physiol. 1997, 115, 1557–1567. [Google Scholar] [CrossRef] [Green Version]
- Kurata, N.; Kariu, T.; Kawano, S.; Kimura, M. Molecular cloning of cDNAs encoding ribonuclease-related proteins in Nicotiana glutinosa leaves, as induced in response to wounding or to TMV-infection. Biosci. Biotechnol. Biochem. 2002, 66, 391–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacIntosh, G.C.; Hillwig, M.S.; Meyer, A.; Flagel, L. RNase T2 genes from rice and the evolution of secretory ribonucleases in plants. Mol. Genet. Genom. 2010, 283, 381–396. [Google Scholar] [CrossRef]
- Baranzini, N.; Monti, L.; Vanotti, M.; Orlandi, V.T.; Bolognese, F.; Scaldaferri, D.; Girardello, R.; Tettamanti, G.; de Eguileor, M.; Vizioli, J.; et al. AIF-1 and RNASET2 Play Complementary Roles in the Innate Immune Response of Medicinal Leech. J. Innate Immun. 2019, 11, 150–167. [Google Scholar] [CrossRef]
- Hugot, K.; Ponchet, M.; Marais, A.; Ricci, P.; Galiana, E. A tobacco S-like RNase inhibits hyphal elongation of plant pathogens. Mol. Plant Microbe Interact. 2002, 15, 243–250. [Google Scholar] [CrossRef] [Green Version]
- Loffler, A.; Abel, S.; Jost, W.; Beintema, J.J.; Glund, K. Phosphate-Regulated Induction of Intracellular Ribonucleases in Cultured Tomato (Lycopersicon esculentum) Cells. Plant Physiol. 1992, 98, 1472–1478. [Google Scholar] [CrossRef] [Green Version]
- Nurnberger, T.; Abel, S.; Jost, W.; Glund, K. Induction of an Extracellular Ribonuclease in Cultured Tomato Cells upon Phosphate Starvation. Plant Physiol. 1990, 92, 970–976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.L.; Delatorre, C.A.; Bakker, A.; Abel, S. Conditional identification of phosphate-starvation-response mutants in Arabidopsis thaliana. Planta 2000, 211, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Tasaki, Y.; Azwan, A.; Hara, T.; Joh, T. Structure and expression of a phosphate deficiency-inducible ribonuclease gene in Pholiota nameko. Curr. Genet. 2004, 45, 28–36. [Google Scholar] [CrossRef]
- McGugan, G.C., Jr.; Joshi, M.B.; Dwyer, D.M. Identification and biochemical characterization of unique secretory nucleases of the human enteric pathogen, Entamoeba histolytica. J. Biol. Chem. 2007, 282, 31789–31802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hillwig, M.S.; Contento, A.L.; Meyer, A.; Ebany, D.; Bassham, D.C.; Macintosh, G.C. RNS2, a conserved member of the RNase T2 family, is necessary for ribosomal RNA decay in plants. Proc. Natl. Acad. Sci. USA 2011, 108, 1093–1098. [Google Scholar] [CrossRef]
- Haud, N.; Kara, F.; Diekmann, S.; Henneke, M.; Willer, J.R.; Hillwig, M.S.; Gregg, R.G.; Macintosh, G.C.; Gartner, J.; Alia, A.; et al. rnaset2 mutant zebrafish model familial cystic leukoencephalopathy and reveal a role for RNase T2 in degrading ribosomal RNA. Proc. Natl. Acad. Sci. USA 2011, 108, 1099–1103. [Google Scholar] [CrossRef]
- Weber, T.; Schlotawa, L.; Dosch, R.; Hamilton, N.; Kaiser, J.; Schiller, S.; Wenske, B.; Gartner, J.; Henneke, M. Zebrafish disease model of human RNASET2-deficient cystic leukoencephalopathy displays abnormalities in early microglia. Biol. Open 2020, 9, bio049239. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, N.; Rutherford, H.A.; Petts, J.J.; Isles, H.M.; Weber, T.; Henneke, M.; Gartner, J.; Dunning, M.J.; Renshaw, S.A. The failure of microglia to digest developmental apoptotic cells contributes to the pathology of RNASET2-deficient leukoencephalopathy. Glia 2020, 68, 1531–1545. [Google Scholar] [CrossRef] [Green Version]
- Kettwig, M.; Ternka, K.; Wendland, K.; Kruger, D.M.; Zampar, S.; Schob, C.; Franz, J.; Aich, A.; Winkler, A.; Sakib, M.S.; et al. Interferon-driven brain phenotype in a mouse model of RNaseT2 deficient leukoencephalopathy. Nat. Commun. 2021, 12, 6530. [Google Scholar] [CrossRef] [PubMed]
- Kameli, R.; Amanat, M.; Rezaei, Z.; Hosseionpour, S.; Nikbakht, S.; Alizadeh, H.; Ashrafi, M.R.; Omrani, A.; Garshasbi, M.; Tavasoli, A.R. RNASET2-deficient leukoencephalopathy mimicking congenital CMV infection and Aicardi-Goutieres syndrome: A case report with a novel pathogenic variant. Orphanet J. Rare Dis. 2019, 14, 184. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruno, A.; Noonan, D.M.; Valli, R.; Porta, G.; Taramelli, R.; Mortara, L.; Acquati, F. Human RNASET2: A Highly Pleiotropic and Evolutionary Conserved Tumor Suppressor Gene Involved in the Control of Ovarian Cancer Pathogenesis. Int. J. Mol. Sci. 2022, 23, 9074. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, X.; Xiang, L. Role and Mechanism of RNASET2 in the Pathogenesis of Vitiligo. J. Investig. Dermatol. Symp. Proc. 2015, 17, 48–50. [Google Scholar] [CrossRef]
- De Vito, A.; Orecchia, P.; Balza, E.; Reverberi, D.; Scaldaferri, D.; Taramelli, R.; Noonan, D.M.; Acquati, F.; Mortara, L. Overexpression of Murine Rnaset2 in a Colon Syngeneic Mouse Carcinoma Model Leads to Rebalance of Intra-Tumor M1/M2 Macrophage Ratio, Activation of T Cells, Delayed Tumor Growth, and Rejection. Cancers 2020, 12, 717. [Google Scholar] [CrossRef] [Green Version]
- Trubia, M.; Sessa, L.; Taramelli, R. Mammalian Rh/T2/S-glycoprotein ribonuclease family genes: Cloning of a human member located in a region of chromosome 6 (6q27) frequently deleted in human malignancies. Genomics 1997, 42, 342–344. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Ragoussis, J.; Stamp, G.W.; Allan, G.J.; Trowsdale, J. Frequent loss of heterozygosity on chromosome 6 in human ovarian carcinoma. Br. J. Cancer 1993, 67, 551–559. [Google Scholar] [CrossRef] [Green Version]
- Queimado, L.; Reis, A.; Fonseca, I.; Martins, C.; Lovett, M.; Soares, J.; Parreira, L. A refined localization of two deleted regions in chromosome 6q associated with salivary gland carcinomas. Oncogene 1998, 16, 83–88. [Google Scholar] [CrossRef] [Green Version]
- Chappell, S.A.; Walsh, T.; Walker, R.A.; Shaw, J.A. Loss of heterozygosity at chromosome 6q in preinvasive and early invasive breast carcinomas. Br. J. Cancer 1997, 75, 1324–1329. [Google Scholar] [CrossRef] [Green Version]
- Queimado, L.; Seruca, R.; Costa-Pereira, A.; Castedo, S. Identification of two distinct regions of deletion at 6q in gastric carcinoma. Genes. Chromosomes Cancer 1995, 14, 28–34. [Google Scholar] [CrossRef]
- Tahara, H.; Smith, A.P.; Gas, R.D.; Cryns, V.L.; Arnold, A. Genomic localization of novel candidate tumor suppressor gene loci in human parathyroid adenomas. Cancer Res. 1996, 56, 599–605. [Google Scholar]
- Millikin, D.; Meese, E.; Vogelstein, B.; Witkowski, C.; Trent, J. Loss of heterozygosity for loci on the long arm of chromosome 6 in human malignant melanoma. Cancer Res. 1991, 51, 5449–5453. [Google Scholar] [PubMed]
- Acquati, F.; Morelli, C.; Cinquetti, R.; Bianchi, M.G.; Porrini, D.; Varesco, L.; Gismondi, V.; Rocchetti, R.; Talevi, S.; Possati, L.; et al. Cloning and characterization of a senescence inducing and class II tumor suppressor gene in ovarian carcinoma at chromosome region 6q27. Oncogene 2001, 20, 980–988. [Google Scholar] [CrossRef] [Green Version]
- Acquati, F.; Possati, L.; Ferrante, L.; Campomenosi, P.; Talevi, S.; Bardelli, S.; Margiotta, C.; Russo, A.; Bortoletto, E.; Rocchetti, R.; et al. Tumor and metastasis suppression by the human RNASET2 gene. Int. J. Oncol. 2005, 26, 1159–1168. [Google Scholar] [CrossRef] [PubMed]
- Yin, F.; Liu, L.; Liu, X.; Li, G.; Zheng, L.; Li, D.; Wang, Q.; Zhang, W.; Li, L. Downregulation of tumor suppressor gene ribonuclease T2 and gametogenetin binding protein 2 is associated with drug resistance in ovarian cancer. Oncol. Rep. 2014, 32, 362–372. [Google Scholar] [CrossRef] [PubMed]
- Roggiani, F.; Riva, C.; Raspagliesi, F.; Porta, G.; Valli, R.; Taramelli, R.; Acquati, F.; Mezzanzanica, D.; Tomassetti, A. A Cell-Autonomous Oncosuppressive Role of Human RNASET2 Affecting ECM-Mediated Oncogenic Signaling. Cancers 2019, 11, 255. [Google Scholar] [CrossRef] [Green Version]
- Ji, M.; Zhao, Z.; Li, Y.; Xu, P.; Shi, J.; Li, Z.; Wang, K.; Huang, X.; Liu, B. FBXO6-mediated RNASET2 ubiquitination and degradation governs the development of ovarian cancer. Cell Death Dis. 2021, 12, 317. [Google Scholar] [CrossRef]
- Roiz, L.; Smirnoff, P.; Bar-Eli, M.; Schwartz, B.; Shoseyov, O. ACTIBIND, an actin-binding fungal T2-RNase with antiangiogenic and anticarcinogenic characteristics. Cancer 2006, 106, 2295–2308. [Google Scholar] [CrossRef] [Green Version]
- Polakowski, N.; Han, H.; Lemasson, I. Direct inhibition of RNAse T2 expression by the HTLV-1 viral protein Tax. Viruses 2011, 3, 1485–1500. [Google Scholar] [CrossRef] [Green Version]
- Wang, Q.; Jiang, M.; Wu, J.; Ma, Y.; Li, T.; Chen, Q.; Zhang, X.; Xiang, L. Stress-induced RNASET2 overexpression mediates melanocyte apoptosis via the TRAF2 pathway in vitro. Cell Death Dis. 2014, 5, e1022. [Google Scholar] [CrossRef] [Green Version]
- Lualdi, M.; Pedrini, E.; Rea, K.; Monti, L.; Scaldaferri, D.; Gariboldi, M.; Camporeale, A.; Ghia, P.; Monti, E.; Tomassetti, A.; et al. Pleiotropic modes of action in tumor cells of RNASET2, an evolutionary highly conserved extracellular RNase. Oncotarget 2015, 6, 7851–7865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Caputa, G.; Zhao, S.; Criado, A.E.; Ory, D.S.; Duncan, J.G.; Schaffer, J.E. RNASET2 is required for ROS propagation during oxidative stress-mediated cell death. Cell Death Differ. 2016, 23, 347–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.; Liu, P.; Zheng, Q.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Tong, T.; et al. Mitochondrial Trafficking and Processing of Telomerase RNA TERC. Cell Rep. 2018, 24, 2589–2595. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Q.; Liu, P.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Di, F.; Tong, T.; et al. Mitochondrion-processed TERC regulates senescence without affecting telomerase activities. Protein Cell 2019, 10, 631–648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smirnoff, P.; Roiz, L.; Angelkovitch, B.; Schwartz, B.; Shoseyov, O. A recombinant human RNASET2 glycoprotein with antitumorigenic and antiangiogenic characteristics: Expression, purification, and characterization. Cancer 2006, 107, 2760–2769. [Google Scholar] [CrossRef]
- Roiz, L.; Smirnoff, P.; Lewin, I.; Shoseyov, O.; Schwartz, B. Human recombinant RNASET2: A potential anti-cancer drug. Oncoscience 2016, 3, 71–84. [Google Scholar] [CrossRef]
- Schwartz, B.; Shoseyov, O.; Melnikova, V.O.; McCarty, M.; Leslie, M.; Roiz, L.; Smirnoff, P.; Hu, G.F.; Lev, D.; Bar-Eli, M. ACTIBIND, a T2 RNase, competes with angiogenin and inhibits human melanoma growth, angiogenesis, and metastasis. Cancer Res. 2007, 67, 5258–5266. [Google Scholar] [CrossRef] [Green Version]
- Everts, B.; Perona-Wright, G.; Smits, H.H.; Hokke, C.H.; van der Ham, A.J.; Fitzsimmons, C.M.; Doenhoff, M.J.; van der Bosch, J.; Mohrs, K.; Haas, H.; et al. Omega-1, a glycoprotein secreted by Schistosoma mansoni eggs, drives Th2 responses. J. Exp. Med. 2009, 206, 1673–1680. [Google Scholar] [CrossRef]
- Steinfelder, S.; Andersen, J.F.; Cannons, J.L.; Feng, C.G.; Joshi, M.; Dwyer, D.; Caspar, P.; Schwartzberg, P.L.; Sher, A.; Jankovic, D. The major component in schistosome eggs responsible for conditioning dendritic cells for Th2 polarization is a T2 ribonuclease (omega-1). J. Exp. Med. 2009, 206, 1681–1690. [Google Scholar] [CrossRef]
- Everts, B.; Hussaarts, L.; Driessen, N.N.; Meevissen, M.H.; Schramm, G.; van der Ham, A.J.; van der Hoeven, B.; Scholzen, T.; Burgdorf, S.; Mohrs, M.; et al. Schistosome-derived omega-1 drives Th2 polarization by suppressing protein synthesis following internalization by the mannose receptor. J. Exp. Med. 2012, 209, 1753–1767. [Google Scholar] [CrossRef]
- Monaci, S.; Coppola, F.; Giuntini, G.; Roncoroni, R.; Acquati, F.; Sozzani, S.; Carraro, F.; Naldini, A. Hypoxia Enhances the Expression of RNASET2 in Human Monocyte-Derived Dendritic Cells: Role of PI3K/AKT Pathway. Int. J. Mol. Sci. 2021, 22, 7564. [Google Scholar] [CrossRef]
- Scaldaferri, D.; Bosi, A.; Fabbri, M.; Pedrini, E.; Inforzato, A.; Valli, R.; Frattini, A.; De Vito, A.; Noonan, D.M.; Taramelli, R.; et al. The human RNASET2 protein affects the polarization pattern of human macrophages in vitro. Immunol. Lett. 2018, 203, 102–111. [Google Scholar] [CrossRef]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef]
- Greulich, W.; Wagner, M.; Gaidt, M.M.; Stafford, C.; Cheng, Y.; Linder, A.; Carell, T.; Hornung, V. TLR8 Is a Sensor of RNase T2 Degradation Products. Cell 2019, 179, 1264–1275.E13. [Google Scholar] [CrossRef] [PubMed]
- Gallo, D.; De Vito, A.; Roncoroni, R.; Bruno, A.; Piantanida, E.; Bartalena, L.; Tanda, M.L.; Mortara, L.; Acquati, F. A potential role of human RNASET2 overexpression in the pathogenesis of Graves’ disease. Endocrine 2023, 79, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Acquati, F.; Bertilaccio, S.; Grimaldi, A.; Monti, L.; Cinquetti, R.; Bonetti, P.; Lualdi, M.; Vidalino, L.; Fabbri, M.; Sacco, M.G.; et al. Microenvironmental control of malignancy exerted by RNASET2, a widely conserved extracellular RNase. Proc. Natl. Acad. Sci. USA 2011, 108, 1104–1109. [Google Scholar] [CrossRef]
- Sica, A.; Larghi, P.; Mancino, A.; Rubino, L.; Porta, C.; Totaro, M.G.; Rimoldi, M.; Biswas, S.K.; Allavena, P.; Mantovani, A. Macrophage polarization in tumour progression. Semin. Cancer Biol. 2008, 18, 349–355. [Google Scholar] [CrossRef]
- Parisi, L.; Gini, E.; Baci, D.; Tremolati, M.; Fanuli, M.; Bassani, B.; Farronato, G.; Bruno, A.; Mortara, L. Macrophage Polarization in Chronic Inflammatory Diseases: Killers or Builders? J. Immunol. Res. 2018, 2018, 8917804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acquati, F.; Lualdi, M.; Bertilaccio, S.; Monti, L.; Turconi, G.; Fabbri, M.; Grimaldi, A.; Anselmo, A.; Inforzato, A.; Collotta, A.; et al. Loss of function of Ribonuclease T2, an ancient and phylogenetically conserved RNase, plays a crucial role in ovarian tumorigenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 8140–8145. [Google Scholar] [CrossRef]
- Zeng, Z.; Zhang, X.; Li, D.; Li, J.; Yuan, J.; Gu, L.; Xiong, X. Expression, Location, Clinical Implication, and Bioinformatics Analysis of RNASET2 in Gastric Adenocarcinoma. Front. Oncol. 2020, 10, 836. [Google Scholar] [CrossRef]
- Uccella, S.; La Rosa, S.; Scaldaferri, D.; Monti, L.; Maragliano, R.; Sorrenti, E.; Gariboldi, M.; Taramelli, R.; Sessa, F.; Acquati, F. New insights into hypoxia-related mechanisms involved in different microvascular patterns of bronchopulmonary carcinoids and poorly differentiated neuroendocrine carcinomas. Role of ribonuclease T2 (RNASET2) and HIF-1alpha. Hum. Pathol. 2018, 79, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Sandler, A.; Gray, R.; Perry, M.C.; Brahmer, J.; Schiller, J.H.; Dowlati, A.; Lilenbaum, R.; Johnson, D.H. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N. Engl. J. Med. 2006, 355, 2542–2550. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Campomenosi, P.; Mortara, L.; Bassani, B.; Valli, R.; Porta, G.; Bruno, A.; Acquati, F. The Potential Role of the T2 Ribonucleases in TME-Based Cancer Therapy. Biomedicines 2023, 11, 2160. https://doi.org/10.3390/biomedicines11082160
Campomenosi P, Mortara L, Bassani B, Valli R, Porta G, Bruno A, Acquati F. The Potential Role of the T2 Ribonucleases in TME-Based Cancer Therapy. Biomedicines. 2023; 11(8):2160. https://doi.org/10.3390/biomedicines11082160
Chicago/Turabian StyleCampomenosi, Paola, Lorenzo Mortara, Barbara Bassani, Roberto Valli, Giovanni Porta, Antonino Bruno, and Francesco Acquati. 2023. "The Potential Role of the T2 Ribonucleases in TME-Based Cancer Therapy" Biomedicines 11, no. 8: 2160. https://doi.org/10.3390/biomedicines11082160
APA StyleCampomenosi, P., Mortara, L., Bassani, B., Valli, R., Porta, G., Bruno, A., & Acquati, F. (2023). The Potential Role of the T2 Ribonucleases in TME-Based Cancer Therapy. Biomedicines, 11(8), 2160. https://doi.org/10.3390/biomedicines11082160