Preliminary Study of the Relationship between Osteopontin and Relapsed Hodgkin’s Lymphoma
Abstract
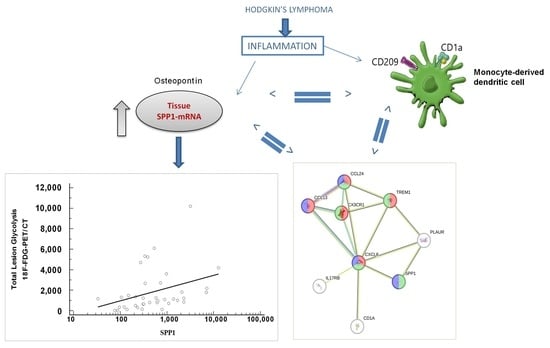
:1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
2.2. Maximum Standardized Uptake Value
2.3. Immunohistochemistry
2.4. RNA Extraction and NanoString Quantification
2.5. Statistical Analysis
3. Results
3.1. SPP1, Clinical Information, Laboratory Results, and Pathological Data
3.2. Higher SPP1 Count Associated with a Poor Prognosis
3.3. Positive Correlation between Value of PET Imaging and SPP1
3.4. Comparing SPP1 to TLG for Predicting Tumor Relapse or Progression
3.5. nCounter Immune Expression Analysis of the SPP1 Signature
3.6. SPP1 Signature Is Associated with Dendritic Immune Cell Types
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Participants in the Italian AIEOP Research Network for Hodgkin’s Lymphoma Include
| Irene D’Alba | Azienda Ospedali Riuniti Presidio “G. Salesi”, Italy |
| Maurizio Mascarin | IRCCS Centro Di Riferimento Oncologico—Aviano, Italy |
| Paola Muggeo | AOU Policlinico, Italy |
| Massimo Provenzi | Ospedale Papa Giovanni XXIII, Italy |
| Rosamaria Mura | Ospedale Pediatrico Microcitemico “Antonio Cao”, Azienda Ospedaliera Brotzu, Italy |
| Eulalia Galea | A.O. Pugliese-Ciaccio, Italy |
| Domenico Sperlì | SO “Annunziata”, Italy |
| Simona Rinieri | AOU Sant’Anna Di Ferrara, Italy |
| Tommaso Casini | Azienda Ospedaliero-Universitaria “Anna Meyer”, Italy |
| Alberto Garaventa | IRCCS “Istituto Giannina Gaslini”, Italy |
| Alessandra Sala | Fondazione MBBM, Italy |
| Salvatore Buffardi | AORN Santobono-Pausilipon, Italy |
| Francesca Rossi | Università Degli Studi della Campania “Luigi Vanvitelli”, Italy |
| Marta Pillon | Azienda Ospedaliera Di Padova, Italy |
| Farruggia | ARNAS Civico Di Cristina E Benfratelli, Italy |
| Patrizia Bertolini | Azienda Ospedaliero Universitaria Di Parma, Italy |
| Katia Perruccio | A.O.U. “S.M. Della Misericordia” di Perugia, Italy |
| Antonella Sau | Ospedale Civile Dello Spirito Santo, Italy |
| Sayla Bernasconi | Azienda Ospedaliero-Universitaria Pisana, Italy |
| Roberta Pericoli | Ospedale Infermi, Italy |
| Luciana Vinti | IRCCS Ospedale Pediatrico Bambino Gesù, Italy |
| Fiorina Giona | “Sapienza” Università di Roma, Italy |
| Raffaella De Santis | IRCCS Ospedale “Casa Sollievo Della Sofferenza”, Italy |
| Maurizio Bianchi | AOU Città Della Salute E Della Scienza Di Torino—Presidio Infantile Regina Margherita, Italy |
| Simone Cesaro | Ospedale Donna Bambino - Azienda Ospedaliera Universitaria Integrata, Italy |
| Elena Facchini | Policlinico Sant’Orsola Malpighi, Italy |
| Salvatore D’Amico | AOU Policlinico Vittorio Emanuele, Italy |
| Valerio Cecinati | Ospedale SS. Annunziata, Italy |
References
- SEER Cancer Statistics Review, 1975–2017. Available online: https://seer.cancer.gov/csr/1975_2017/index.html (accessed on 8 August 2023).
- AIRTUM Working Group; CCM; AIEOP Working Group. Italian Cancer Figures, Report 2012: Cancer in Children and Adolescents. Epidemiol. Prev. 2013, 37, 1–225. [Google Scholar]
- Burnelli, R.; Fiumana, G.; Rondelli, R.; Pillon, M.; Sala, A.; Garaventa, A.; D’Amore, E.S.G.; Sabattini, E.; Buffardi, S.; Bianchi, M.; et al. Comparison of Hodgkin’s Lymphoma in Children and Adolescents. A Twenty Year Experience with MH’96 and LH2004 AIEOP (Italian Association of Pediatric Hematology and Oncology) Protocols. Cancers 2020, 12, 1620. [Google Scholar] [CrossRef] [PubMed]
- De Re, V.; Repetto, O.; Mussolin, L.; Brisotto, G.; Elia, C.; Lopci, E.; d’Amore, E.S.G.; Burnelli, R.; Mascarin, M. Promising Drugs and Treatment Options for Pediatric and Adolescent Patients with Hodgkin Lymphoma. Front. Cell Dev. Biol. 2022, 10, 965803. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D.; Tsang, D.S.; Tinkle, C.L.; Olch, A.J.; Kremer, L.C.M.; Ronckers, C.M.; Gibbs, I.C.; Constine, L.S. Late Effects of Radiation Therapy in Pediatric Patients and Survivorship. Pediatr. Blood Cancer 2021, 68, e28349. [Google Scholar] [CrossRef]
- Second International Inter-Group Study for Classical Hodgkin Lymphoma in Children and Adolescents. Available online: https://Clinicaltrials.Gov/Ct2/Show/NCT02684708 (accessed on 13 May 2021).
- Lopci, E.; Burnelli, R.; Elia, C.; Piccardo, A.; Castello, A.; Borsatti, E.; Zucchetta, P.; Cistaro, A.; Mascarin, M.; AIEOP Hodgkin Lymphoma Study Group. Additional Value of Volumetric and Texture Analysis on FDG PET Assessment in Paediatric Hodgkin Lymphoma: An Italian Multicentric Study Protocol. BMJ Open 2021, 11, e041252. [Google Scholar] [CrossRef]
- Abdel-Haleem, A.M.; Lewis, N.E.; Jamshidi, N.; Mineta, K.; Gao, X.; Gojobori, T. The Emerging Facets of Non-Cancerous Warburg Effect. Front. Endocrinol. 2017, 8, 279. [Google Scholar] [CrossRef]
- Kluge, R.; Chavdarova, L.; Hoffmann, M.; Kobe, C.; Malkowski, B.; Montravers, F.; Kurch, L.; Georgi, T.; Dietlein, M.; Wallace, W.H.; et al. Inter-Reader Reliability of Early FDG-PET/CT Response Assessment Using the Deauville Scale after 2 Cycles of Intensive Chemotherapy (OEPA) in Hodgkin’s Lymphoma. PLoS ONE 2016, 11, e0149072. [Google Scholar] [CrossRef]
- Akhtari, M.; Milgrom, S.A.; Pinnix, C.C.; Reddy, J.P.; Dong, W.; Smith, G.L.; Mawlawi, O.; Abou Yehia, Z.; Gunther, J.; Osborne, E.M.; et al. Reclassifying Patients with Early-Stage Hodgkin Lymphoma Based on Functional Radiographic Markers at Presentation. Blood 2018, 131, 84–94. [Google Scholar] [CrossRef]
- Lopci, E.; Elia, C.; Catalfamo, B.; Burnelli, R.; De Re, V.; Mussolin, L.; Piccardo, A.; Cistaro, A.; Borsatti, E.; Zucchetta, P.; et al. Prospective Evaluation of Different Methods for Volumetric Analysis on [18F]FDG PET/CT in Pediatric Hodgkin Lymphoma. J. Clin. Med. 2022, 11, 6223. [Google Scholar] [CrossRef]
- Tan, Y.; Zhao, L.; Yang, Y.-G.; Liu, W. The Role of Osteopontin in Tumor Progression through Tumor-Associated Macrophages. Front. Oncol. 2022, 12, 953283. [Google Scholar] [CrossRef]
- Lovisa, F.; Garbin, A.; Crotti, S.; Di Battista, P.; Gallingani, I.; Damanti, C.C.; Tosato, A.; Carraro, E.; Pillon, M.; Mafakheri, E.; et al. Increased Tenascin C, Osteopontin and HSP90 Levels in Plasmatic Small Extracellular Vesicles of Pediatric ALK-Positive Anaplastic Large Cell Lymphoma: New Prognostic Biomarkers? Diagnostics 2021, 11, 253. [Google Scholar] [CrossRef] [PubMed]
- Pamuk, G.E.; Uyanik, M.S.; Pamuk, O.N.; Maden, M.; Tapan, U. Decreased Dickkopf-1 Levels in Chronic Lymphocytic Leukemia and Increased Osteopontin Levels in Non-Hodgkin’s Lymphoma at Initial Diagnosis: Could They Be Playing Roles in Pathogenesis? Hematol. Amst. Neth. 2015, 20, 267–271. [Google Scholar] [CrossRef] [PubMed]
- Duletić-Načinović, A.; Gačić, V.; Valković, T.; Lučin, K.; Fišić, E.; Žuvić-Butorac, M.; Seili-Bekafigo, I.; Jonjić, N. Concurrent Elevations of VEGF, Osteopontin and MCP-1 Serum Levels Are Independent Predictors of Survival in Patients with Diffuse Large B-Cell Lymphoma. Acta Haematol. 2016, 136, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ye, G.; Dong, B.; Huang, L.; Zhang, C.; Sheng, Y.; Wu, B.; Han, L.; Wu, C.; Qi, Y. A Pan-Cancer Analysis of the Oncogenic Role of Secreted Phosphoprotein 1 (SPP1) in Human Cancers. Ann. Transl. Med. 2022, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yin, X.S.; Guo, H.; Han, R.K.; He, R.D.; Chi, L.J. Elevated Osteopontin Levels in Mild Cognitive Impairment and Alzheimer’s Disease. Mediators Inflamm. 2013, 2013, 615745. [Google Scholar] [CrossRef] [PubMed]
- Rittling, S.R.; Singh, R. Osteopontin in Immune-Mediated Diseases. J. Dent. Res. 2015, 94, 1638–1645. [Google Scholar] [CrossRef] [PubMed]
- Clemente, N.; Raineri, D.; Cappellano, G.; Boggio, E.; Favero, F.; Soluri, M.F.; Dianzani, C.; Comi, C.; Dianzani, U.; Chiocchetti, A. Osteopontin Bridging Innate and Adaptive Immunity in Autoimmune Diseases. J. Immunol. Res. 2016, 2016, 7675437. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Morser, J.; Leung, L.L.K. Thrombin Cleavage of Osteopontin Disrupts a Pro-Chemotactic Sequence for Dendritic Cells, Which Is Compensated by the Release of Its Pro-Chemotactic C-Terminal Fragment. J. Biol. Chem. 2014, 289, 27146–27158. [Google Scholar] [CrossRef]
- Yamaguchi, Y.; Shao, Z.; Sharif, S.; Du, X.-Y.; Myles, T.; Merchant, M.; Harsh, G.; Glantz, M.; Recht, L.; Morser, J.; et al. Thrombin-Cleaved Fragments of Osteopontin Are Overexpressed in Malignant Glial Tumors and Provide a Molecular Niche with Survival Advantage. J. Biol. Chem. 2013, 288, 3097–3111. [Google Scholar] [CrossRef]
- Peraramelli, S.; Zhou, Q.; Zhou, Q.; Wanko, B.; Zhao, L.; Nishimura, T.; Leung, T.H.; Mizuno, S.; Ito, M.; Myles, T.; et al. Thrombin Cleavage of Osteopontin Initiates Osteopontin’s Tumor-Promoting Activity. J. Thromb. Haemost. JTH 2022, 20, 1256–1270. [Google Scholar] [CrossRef]
- Leung, L.L.; Myles, T.; Morser, J. Thrombin Cleavage of Osteopontin and the Host Anti-Tumor Immune Response. Cancers 2023, 15, 3480. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Mirza, M.; Weber, G.F. An Osteopontin Splice Variant Induces Anchorage Independence in Human Breast Cancer Cells. Oncogene 2006, 25, 2192–2202. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Gu, K.; He, J.; Sharma, S. Preferential Up-Regulation of Osteopontin in Primary Central Nervous System Lymphoma Does Not Correlate with Putative Receptor CD44v6 or CD44H Expression. Hum. Pathol. 2013, 44, 606–611. [Google Scholar] [CrossRef] [PubMed]
- Soldan, S.S.; Su, C.; Lamontagne, R.J.; Grams, N.; Lu, F.; Zhang, Y.; Gesualdi, J.D.; Frase, D.M.; Tolvinski, L.E.; Martin, K.; et al. Epigenetic Plasticity Enables CNS-Trafficking of EBV-Infected B Lymphocytes. PLoS Pathog. 2021, 17, e1009618. [Google Scholar] [CrossRef] [PubMed]
- Han, B.; Huang, J.; Han, Y.; Hao, J.; Wu, X.; Song, H.; Chen, X.; Shen, Q.; Dong, X.; Pang, H.; et al. The microRNA miR-181c Enhances Chemosensitivity and Reduces Chemoresistance in Breast Cancer Cells via down-Regulating Osteopontin. Int. J. Biol. Macromol. 2019, 125, 544–556. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Wei, L.; Zhao, W.; Wang, X.; Zheng, G.; Zheng, M.; Song, X.; Zuo, W. Down-Regulation of Osteopontin Expression by RNA Interference Affects Cell Proliferation and Chemotherapy Sensitivity of Breast Cancer MDA-MB-231 Cells. Mol. Med. Rep. 2012, 5, 373–376. [Google Scholar] [CrossRef]
- Ghanbarnasab Behbahani, R.; Danyaei, A.; Teimoori, A.; Tahmasbi, M.J.; Neisi, N. CRISPR/Cas9 Mediated Knocking out of OPN Gene Enhances Radiosensitivity in MDA-MB-231 Breast Cancer Cell Line. J. Cancer Res. Clin. Oncol. 2023, 149, 4117–4130. [Google Scholar] [CrossRef]
- Wu, H.-H.; Hwang-Verslues, W.W.; Lee, W.-H.; Huang, C.-K.; Wei, P.-C.; Chen, C.-L.; Shew, J.-Y.; Lee, E.Y.-H.P.; Jeng, Y.-M.; Tien, Y.-W.; et al. Targeting IL-17B–IL-17RB Signaling with an Anti–IL-17RB Antibody Blocks Pancreatic Cancer Metastasis by Silencing Multiple Chemokines. J. Exp. Med. 2015, 212, 333–349. [Google Scholar] [CrossRef]
- Alinejad, V.; Hossein Somi, M.; Baradaran, B.; Akbarzadeh, P.; Atyabi, F.; Kazerooni, H.; Samadi Kafil, H.; Aghebati Maleki, L.; Siah Mansouri, H.; Yousefi, M. Co-Delivery of IL17RB siRNA and Doxorubicin by Chitosan-Based Nanoparticles for Enhanced Anticancer Efficacy in Breast Cancer Cells. Biomed. Pharmacother. Biomed. Pharmacother. 2016, 83, 229–240. [Google Scholar] [CrossRef]
- Madunić, J. The Urokinase Plasminogen Activator System in Human Cancers: An Overview of Its Prognostic and Predictive Role. Thromb. Haemost. 2018, 118, 2020–2036. [Google Scholar] [CrossRef]
- Repetto, O.; De Re, V.; Mussolin, L.; Tedeschi, M.; Elia, C.; Bianchi, M.; Buffardi, S.; Sala, A.; Burnelli, R.; Mascarin, M. Proteomic Profiles and Biological Processes of Relapsed vs. Non-Relapsed Pediatric Hodgkin Lymphoma. Int. J. Mol. Sci. 2020, 21, 2185. [Google Scholar] [CrossRef] [PubMed]
- Repetto, O.; Caggiari, L.; De Zorzi, M.; Elia, C.; Mussolin, L.; Buffardi, S.; Pillon, M.; Muggeo, P.; Casini, T.; Steffan, A.; et al. Quantitative Plasma Proteomics to Identify Candidate Biomarkers of Relapse in Pediatric/Adolescent Hodgkin Lymphoma. Int. J. Mol. Sci. 2022, 23, 9911. [Google Scholar] [CrossRef] [PubMed]
- Repetto, O.; De Re, V. Coagulation and Fibrinolysis in Gastric Cancer. Ann. N. Y. Acad. Sci. 2017, 1404, 27–48. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Dai, F.; Wang, L.; Sun, Y.; Mei, L.; Ran, Y.; Ye, F. CCL13 and Human Diseases. Front. Immunol. 2023, 14, 1176639. [Google Scholar] [CrossRef] [PubMed]
- Agrusa, J.E.; Scull, B.P.; Abhyankar, H.A.; Lin, H.; Ozuah, N.W.; Chakraborty, R.; Eckstein, O.S.; Gulati, N.; Fattah, E.A.; El-Mallawany, N.K.; et al. Defining the Inflammatory Plasma Proteome in Pediatric Hodgkin Lymphoma. Cancers 2020, 12, 3603. [Google Scholar] [CrossRef] [PubMed]
- Maggio, E.M.; Van Den Berg, A.; Visser, L.; Diepstra, A.; Kluiver, J.; Emmens, R.; Poppema, S. Common and Differential Chemokine Expression Patterns in Rs Cells of NLP, EBV Positive and Negative Classical Hodgkin Lymphomas. Int. J. Cancer 2002, 99, 665–672. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The Chemokine System in Diverse Forms of Macrophage Activation and Polarization. Trends Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Wang, Y.; Hong, T.; Cheng, B.; Gan, S.; Chen, L.; Zhang, J.; Zuo, L.; Li, J.; Cui, X. Blocking the Autocrine Regulatory Loop of Gankyrin/STAT3/CCL24/CCR3 Impairs the Progression and Pazopanib Resistance of Clear Cell Renal Cell Carcinoma. Cell Death Dis. 2020, 11, 117. [Google Scholar] [CrossRef]
- Eide, H.A.; Knudtsen, I.S.; Sandhu, V.; Løndalen, A.M.; Halvorsen, A.R.; Abravan, A.; Kure, E.H.; Bogsrud, T.V.; Brustugun, O.T.; Kyte, J.A.; et al. Serum Cytokine Profiles and Metabolic Tumor Burden in Patients with Non-Small Cell Lung Cancer Undergoing Palliative Thoracic Radiation Therapy. Adv. Radiat. Oncol. 2018, 3, 130–138. [Google Scholar] [CrossRef]
- Colonna, M. TREMs in the Immune System and Beyond. Nat. Rev. Immunol. 2003, 3, 445–453. [Google Scholar] [CrossRef]
- Muller, M.; Haghnejad, V.; Lopez, A.; Tiotiu, A.; Renaud, S.; Derive, M.; Bronowicki, J.-P. Triggering Receptors Expressed on Myeloid Cells 1: Our New Partner in Human Oncology? Front. Oncol. 2022, 12, 927440. [Google Scholar] [CrossRef] [PubMed]
- Gerhard, G.M.; Bill, R.; Messemaker, M.; Klein, A.M.; Pittet, M.J. Tumor-Infiltrating Dendritic Cell States Are Conserved across Solid Human Cancers. J. Exp. Med. 2021, 218, e20200264. [Google Scholar] [CrossRef] [PubMed]
- Chow, K.V.; Lew, A.M.; Sutherland, R.M.; Zhan, Y. Monocyte-Derived Dendritic Cells Promote Th Polarization, Whereas Conventional Dendritic Cells Promote Th Proliferation. J. Immunol. 2016, 196, 624–636. [Google Scholar] [CrossRef]
- Wegrecki, M. CD1a-Mediated Immunity from a Molecular Perspective. Mol. Immunol. 2023, 158, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Cernadas, M.; Lu, J.; Watts, G.; Brenner, M.B. CD1a Expression Defines an Interleukin-12 Producing Population of Human Dendritic Cells. Clin. Exp. Immunol. 2009, 155, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Geijtenbeek, T.B.; Torensma, R.; van Vliet, S.J.; van Duijnhoven, G.C.; Adema, G.J.; van Kooyk, Y.; Figdor, C.G. Identification of DC-SIGN, a Novel Dendritic Cell-Specific ICAM-3 Receptor That Supports Primary Immune Responses. Cell 2000, 100, 575–585. [Google Scholar] [CrossRef]
- Marzaioli, V.; Canavan, M.; Floudas, A.; Flynn, K.; Mullan, R.; Veale, D.J.; Fearon, U. CD209/CD14+ Dendritic Cells Characterization in Rheumatoid and Psoriatic Arthritis Patients: Activation, Synovial Infiltration, and Therapeutic Targeting. Front. Immunol. 2022, 12, 722349. [Google Scholar] [CrossRef]










| SPP1Low | SPP1High | p-Value * | |
|---|---|---|---|
| Clinical data | |||
| Age (mean) | 15 | 14 | 0.358 |
| Sex (M/F) | 15/18 | 5/6 | 1.000 |
| Stage (I-II/III-IV) | 7/26 | 3/8 | 0.681 |
| B-symptom (A/B) | 10/23 | 3/8 | 0.850 |
| Bulky (yes/no) 1nv | 16/16 | 5/6 | 0.797 |
| Extranodal disease (yes/no) | 20/13 | 5/6 | 0.385 |
| Pleura (yes/no) 26 nv | 7/6 | 4/1 | 0.322 |
| Lung (yes/no) 26 nv | 9/4 | 3/2 | 0.718 |
| Pericardic (yes/no) 26 nv | 0/13 | 0/5 | -- |
| Liver (yes/no) 26 nv | 2/11 | 0/5 | 0.366 |
| Bone (yes/no) 26 nv | 6/7 | 0/5 | 0.114 |
| bone marrow (yes/no) 26 nv | 5/8 | 2/3 | 0.954 |
| Therapeutic group (1–2/3) | 9/24 | 2/9 | 0.580 |
| Relapse/Progression (yes/no) | 9/24 | 5/6 | 0.268 |
| Early response (adequate response/inadequate response or progression) | 24/9 | 6/5 | 0.268 |
| Late response (adequate response/inadequate response or progression) | 7/2 | 2/3 | 0.265 |
| Deauville (>2/≤2) | 4/5 | 4/1 | 0.301 |
| Laboratory data | |||
| Erythrocyte sedimentation rate (median) | 84 | 19 | 0.109 |
| Albumin (median) | 3.8 | 3.3 | 0.052 |
| Alkaline phosphatase (median) 25 nv | 127 | 124 | 0.351 |
| C-reactive protein (median) | 11 | 9 | 0.949 |
| Ferritin (median) 10 nv | 256 | 335 | 0.792 |
| Hemoglobin (median) 6 nv | 10.7 | 9.5 | 0.073 |
| Immunoglobulin A (median) | 261 | 201 | 0.298 |
| Immunoglobulin G (median) | 1320 | 1462 | 0.412 |
| Immunoglobulin M (median) | 101 | 136 | 0.891 |
| Lactate dehydrogenase (median) 6 nv | 360 | 264 | 0.786 |
| Lymphocytes | 1.4 | 1.2 | 0.851 |
| White blood cell | 10 | 13 | 0.160 |
| Neutrophils | 10 | 9 | 0.866 |
| Platelets | 358 | 437 | 0.427 |
| Fibrinogen 5 nv | 477 | 574 | 0.228 |
| Protein | 7.4 | 7.7 | 0.355 |
| Histological data | |||
| Histotype (Nodular Sclerosis/Mixed cellularity) 2 others | 20/6 | 11/7 | 0.720 |
| Necrosis (yes/no) 14 nv | 2/19 | 7/2 | 0.042 |
| Inflammation (yes/no) 14 nv | 4/17 | 5/4 | 0.049 |
| Grading (1/2) 14 nv | 5/16 | 5/4 | 0.119 |
| CD15 (+/−) 25 nv | 8/2 | 7/2 | 0.386 |
| CD20 (+/−) 4 nv | 5/24 | 2/9 | 0.945 |
| CD3 (+/−) 15 nv | 0 | 0 | --- |
| CD30 (+/−) 4nv | 29 | 11 | --- |
| PAX5 (+/−) 4 nv | 26/3 | 8/3 | 0.186 |
| OCT2 (+/−) 2 nv | 4/16 | 1/8 | 0.565 |
| CD163 (+/−) 12 nv | 7/16 | 6/3 | 0.147 |
| CD204 (+/−) 16 nv | 1/19 | 1/7 | 0.494 |
| CD11c/CD163 (−1/1) 15 nv | 16/4 | 6/3 | 0.642 |
| CD68r_7% 15 nv | 10/10 | 8/1 | 0.096 |
| Epstein–Barr virus (positive/negative) | 8/25 | 4/7 | 0.441 |
| Gene | Gene Description | Log2 FC | Lower CL | Upper CL | p-Value | Bonferroni Adjusted p-Value |
|---|---|---|---|---|---|---|
| IL17RB | Interleukin-17 receptor B | 1.59 | 1.18 | 2 | 2.86 × 10−9 | 1.73 × 10−6 |
| CCL13 | C-C motif chemokine 13 | 1.5 | 0.872 | 2.13 | 3.08 × 10−5 | 0.0186 |
| CCL24 | C-C motif chemokine 24 | 1.35 | 0.758 | 1.95 | 6.38 × 10−5 | 0.0386 |
| IL8 | Interleukin-8 | 1.21 | 0.722 | 1.69 | 1.65 × 10−5 | 0.00998 |
| F13A1 | Coagulation factor XIII A chain | 1.21 | 0.618 | 1.8 | 0.00025 | 0.151 |
| CCL23 | C-C Motif Chemokine Ligand 23 | 1.2 | 0.654 | 1.74 | 9.90 × 10−5 | 0.0599 |
| CXCL6 | C-X-C Motif Chemokine Ligand 6 | 1.18 | 0.624 | 1.74 | 0.000158 | 0.0957 |
| CCL26 | C-C Motif Chemokine Ligand 26 | 1.15 | 0.573 | 1.72 | 0.000331 | 0.2 |
| CCL11 | C-C Motif Chemokine Ligand 11 | 1.12 | 0.491 | 1.75 | 0.00119 | 0.719 |
| FN1 | Fibronectin 1 | 1.12 | 0.482 | 1.76 | 0.00137 | 0.829 |
| TREM1 | Triggering receptor expressed on myeloid cells | 1.03 | 0.6 | 1.47 | 3.23 × 10−5 | 0.0195 |
| MME | Membrane Metalloendopeptidase | 1.03 | 0.492 | 1.56 | 0.000525 | 0.318 |
| CD209 | C-type lectin receptor | 0.958 | 0.463 | 1.45 | 0.000484 | 0.293 |
| CD1A | T-cell surface glycoprotein CD1a | 0.73 | 0.435 | 1.03 | 1.93 × 10−5 | 0.0117 |
| PLAUR | Urokinase plasminogen activator surface receptor | 0.675 | 0.438 | 0.911 | 1.64 × 10−6 | 0.000995 |
| IL1RN | Interleukin 1 Receptor Antagonist | 0.646 | 0.297 | 0.995 | 0.000794 | 0.48 |
| COL3A1 | Collagen Type III Alpha 1 Chain | 0.615 | 0.265 | 0.965 | 0.00132 | 0.8 |
| CD1E | T-Cell Surface Glycoprotein CD1e | 0.6 | 0.286 | 0.915 | 0.00058 | 0.351 |
| ITGB3 | Integrin Subunit Beta 3 | 0.582 | 0.247 | 0.917 | 0.00151 | 0.915 |
| OSM | Oncostatin M | 0.581 | 0.288 | 0.874 | 0.000377 | 0.228 |
| TNFRSF8 | TNF Receptor Superfamily Member 8 | 0.565 | 0.276 | 0.855 | 0.000433 | 0.262 |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | 0.56 | 0.294 | 0.826 | 0.000174 | 0.105 |
| CXCL3 | Chemokine (C-X-C motif) ligand 3 | 0.559 | 0.27 | 0.848 | 0.00049 | 0.296 |
| IL1R2 | Interleukin 1 Receptor Type 2 | 0.559 | 0.241 | 0.877 | 0.00132 | 0.801 |
| TNFRSF12A | TNF Receptor Superfamily Member 12A | 0.547 | 0.243 | 0.851 | 0.00107 | 0.65 |
| LILRA5 | Leukocyte Immunoglobulin-Like Receptor A5 | 0.527 | 0.235 | 0.819 | 0.00102 | 0.614 |
| PLAU | Plasminogen Activator, Urokinase | 0.522 | 0.265 | 0.779 | 0.000277 | 0.168 |
| SELE | Selectin E | 0.5 | 0.272 | 0.729 | 0.000109 | 0.0661 |
| CD276 | Cluster of Differentiation 276 | 0.354 | 0.172 | 0.536 | 0.000447 | 0.271 |
| CX3CR1 | CX3C chemokine receptor 1 | −0.497 | −0.753 | −0.241 | 0.000457 | 0.277 |
| Plasmacytoid DC | Conventional DC Type1 | Conventional DC Type2 | Monocyte-Derived DC | ||||
|---|---|---|---|---|---|---|---|
| CCR7 | C-C chemokine receptor type 7 | CD8A | T-cell surface glycoprotein CD8 alpha chain | CD14 | Monocyte differentiation antigen CD14 | CD14 | Monocyte differentiation antigen CD14 |
| CD209 | CD209 antigen, Pathogen-recognition receptor | ITGAE | Integrin alpha-E heavy chain | CD163 | Scavenger receptor cysteine-rich type 1 protein M130 | CD1A | T-cell surface glycoprotein CD1a |
| CLEC4C | C-type lectin domain family 4 member C | ITGAX | Integrin alpha-X | ITGAM | Integrin alpha-M | CD1C | T-cell surface glycoprotein CD1c |
| NRP1 | Neuropilin-1 | THBD | Thrombomodulin | CX3CR1 | CX3C chemokine receptor 1 | CD209 | CD209 antigen; Pathogen-recognition receptor |
| SIGLEC | Sialic acid-binding immunoglobulin-like lectin | XCR1 | Chemokine XC receptor 1 | CD1C | T-cell surface glycoprotein CD1c | ITAGX | Integrin alpha-X |
| IFNA1 | Interferon alpha-1 | BATF | Basic leucine zipper transcriptional factor ATF-like | CD2 | T-cell surface antigen CD2 | ITGAM | Integrin alpha-M |
| CD4 | T-cell surface glycoprotein CD4 | IRF8 | Interferon regulatory factor 8 | ITAGX | Integrin alpha-X | MRC1 | Macrophage mannose receptor 1 |
| IRF7 | Interferon regulatory factor 7 | IRF4 | Interferon regulatory factor 4 | CD33 | Myeloid cell surface antigen CD33 | IRF4 | Interferon regulatory factor 4 |
| IRF8 | Interferon regulatory factor 8 | CD33 | Myeloid cell surface antigen CD33 | FCERI | High-affinity immunoglobulin epsilon receptor subunit alpha | FCGR1 | High-affinity immunoglobulin gamma Fc receptor I |
| TLR7 | Toll-like receptor 7 | CX3CR1 | CX3C chemokine receptor 1 | FCER1G | High-affinity immunoglobulin epsilon receptor subunit gamma | ||
| TRL9 | Toll-like receptor 9 | IL23 | Interleukin-23 receptor | ||||
| IFNA | Interferon alpha | TNF | Tumor necrosis factor | ||||
| IL6 | Interleukin-6 | ||||||
| TNF | Tumor necrosis factor | ||||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Re, V.; Lopci, E.; Brisotto, G.; Elia, C.; Mussolin, L.; Mascarin, M.; d’Amore, E.S.G.; AIEOP The Hodgkin’s Lymphoma Research Network. Preliminary Study of the Relationship between Osteopontin and Relapsed Hodgkin’s Lymphoma. Biomedicines 2024, 12, 31. https://doi.org/10.3390/biomedicines12010031
De Re V, Lopci E, Brisotto G, Elia C, Mussolin L, Mascarin M, d’Amore ESG, AIEOP The Hodgkin’s Lymphoma Research Network. Preliminary Study of the Relationship between Osteopontin and Relapsed Hodgkin’s Lymphoma. Biomedicines. 2024; 12(1):31. https://doi.org/10.3390/biomedicines12010031
Chicago/Turabian StyleDe Re, Valli, Egesta Lopci, Giulia Brisotto, Caterina Elia, Lara Mussolin, Maurizio Mascarin, Emanuele Stefano Giovanni d’Amore, and AIEOP The Hodgkin’s Lymphoma Research Network. 2024. "Preliminary Study of the Relationship between Osteopontin and Relapsed Hodgkin’s Lymphoma" Biomedicines 12, no. 1: 31. https://doi.org/10.3390/biomedicines12010031
APA StyleDe Re, V., Lopci, E., Brisotto, G., Elia, C., Mussolin, L., Mascarin, M., d’Amore, E. S. G., & AIEOP The Hodgkin’s Lymphoma Research Network. (2024). Preliminary Study of the Relationship between Osteopontin and Relapsed Hodgkin’s Lymphoma. Biomedicines, 12(1), 31. https://doi.org/10.3390/biomedicines12010031