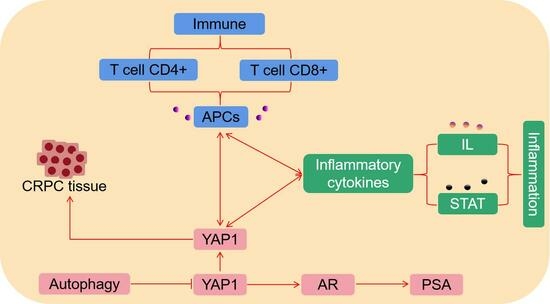
YAP1 Regulates the YAP1/AR/PSA Axis through Autophagy in Castration-Resistant Prostate Cancer and Mediates T-Cell Immune and Inflammatory Cytokine Infiltration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Cultures
2.2. Patients and Tissue Specimens
2.3. Western Blot (WB)
2.4. RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR (qRT-PCR)
2.5. Immunofluorescence Staining
2.6. MTT Assay
2.7. Clone Formation Assay
2.8. Transwell Migration
2.9. Tumor Xenograft Mouse Models
2.10. Statistical Analysis
3. Results
3.1. Distributions and Expressions of YAP1 and PSA Proteins in Different CRPC Tissues and Cell Lines
3.2. YAP1 Promotes the Progression of CRPC Both In Vivo and In Vitro
3.3. Autophagy Regulates the YAP1/AR/PSA Axis in CRPC
3.4. Correlations of YAP1 with the Proportion of Tumor-Infiltrating Immune Cells and Inflammatory Cytokines in PC
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gillessen, S.; Bossi, A.; Davis, I.D.; de Bono, J.; Fizazi, K.; James, N.D.; Mottet, N.; Shore, N.; Small, E.; Smith, M.; et al. Management of Patients with Advanced Prostate Cancer. Part I: Intermediate-/High-risk and Locally Advanced Disease, Biochemical Relapse, and Side Effects of Hormonal Treatment: Report of the Advanced Prostate Cancer Consensus Conference 2022. Eur. Urol. 2023, 83, 267–293. [Google Scholar] [CrossRef] [PubMed]
- Shafi, A.A.; Yen, A.E.; Weigel, N.L. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol. Ther. 2013, 140, 223–238. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Wang, J.; Xu, P.; Zhang, K.; Xin, Z.; Zhao, H.; Ji, Z.; Zhang, M.; Wang, D.; He, Y.; et al. Gremlin1 is a therapeutically targetable FGFR1 ligand that regulates lineage plasticity and castration resistance in prostate cancer. Nat. Cancer 2022, 3, 565–580. [Google Scholar] [CrossRef] [PubMed]
- Small, E.; Saad, F.; Chowdhury, S.; Oudard, S.; Hadaschik, B.; Graff, J.; Olmos, D.; Mainwaring, P.; Lee, J.; Uemura, H.; et al. Apalutamide and overall survival in non-metastatic castration-resistant prostate cancer. Ann. Oncol. 2019, 30, 1813–1820. [Google Scholar] [CrossRef]
- Xiao, L.; Parolia, A.; Qiao, Y.; Bawa, P.; Eyunni, S.; Mannan, R.; Carson, S.E.; Chang, Y.; Wang, X.; Zhang, Y.; et al. Targeting SWI/SNF ATPases in enhancer-addicted prostate cancer. Nature 2022, 601, 434–439. [Google Scholar] [CrossRef]
- Wu, N.; Wang, Y.; Wang, K.; Zhong, B.; Liao, Y.; Liang, J.; Jiang, N. Cathepsin K regulates the tumor growth and metastasis by IL-17/CTSK/EMT axis and mediates M2 macrophage polarization in castration-resistant prostate cancer. Cell Death Dis. 2022, 13, 813. [Google Scholar] [CrossRef]
- Liang, J.; Liao, Y.; Wang, P.; Yang, K.; Wang, Y.; Wang, K.; Zhong, B.; Zhou, D.; Cao, Q.; Li, J.; et al. Ferroptosis landscape in prostate cancer from molecular and metabolic perspective. Cell Death Discov. 2023, 9, 128. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, N.; Jiang, N. Autophagy provides a conceptual therapeutic framework for bone metastasis from prostate cancer. Cell Death Dis. 2021, 12, 909, Corrected in Cell Death Dis. 2021, 12, 1021. [Google Scholar] [CrossRef] [PubMed]
- Mukherji, D.; Omlin, A.; Pezaro, C.; Shamseddine, A.; de Bono, J. Metastatic castration-resistant prostate cancer (CRPC): Preclinical and clinical evidence for the sequential use of novel therapeutics. Cancer Metastasis Rev. 2014, 33, 555–566. [Google Scholar] [CrossRef]
- Moya, I.M.; Halder, G. Hippo-YAP/TAZ signalling in organ regeneration and regenerative medicine. Nat. Rev. Mol. Cell Biol. 2019, 20, 211–226. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, M.; Hu, Y.; Zhao, E.; Li, J.; Ren, L.; Wang, M.; Xu, Y.; Liang, Q.; Zhang, D.; et al. MYBL2 disrupts the Hippo-YAP pathway and confers castration resistance and metastatic potential in prostate cancer. Theranostics 2021, 11, 5794–5812. [Google Scholar] [CrossRef]
- Affo, S.; Nair, A.; Brundu, F.; Ravichandra, A.; Bhattacharjee, S.; Matsuda, M.; Chin, L.; Filliol, A.; Wen, W.; Song, X.; et al. Promotion of cholangiocarcinoma growth by diverse cancer-associated fibroblast subpopulations. Cancer Cell. 2021, 39, 866–882.e11, Corrected in Cancer Cell. 2021, 39, 883. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, S.; Chen, X.; Stauffer, S.; Yu, F.; Lele, S.M.; Fu, K.; Datta, K.; Palermo, N.; Chen, Y.; et al. The hippo pathway effector YAP regulates motility, invasion, and castration-resistant growth of prostate cancer cells. Mol. Cell. Biol. 2015, 35, 1350–1362. [Google Scholar] [CrossRef] [PubMed]
- Glick, D.; Barth, S.; Macleod, K.F. Autophagy: Cellular and molecular mechanisms. J. Pathol. 2010, 221, 3–12. [Google Scholar] [CrossRef]
- White, E. The role for autophagy in cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef]
- Hu, F.; Song, D.; Yan, Y.; Huang, C.; Shen, C.; Lan, J.; Chen, Y.; Liu, A.; Wu, Q.; Sun, L.; et al. IL-6 regulates autophagy and chemotherapy resistance by promoting BECN1 phosphorylation. Nat. Commun. 2021, 12, 3651. [Google Scholar] [CrossRef]
- Xue, J.F.; Shi, Z.M.; Zou, J.; Li, X.L. Inhibition of PI3K/AKT/mTOR signaling pathway promotes autophagy of articular chondrocytes and attenuates inflammatory response in rats with osteoarthritis. Biomed. Pharmacother. 2017, 89, 1252–1261. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Pérez, J.; Carrasco-Rozas, A.; Borrell-Pages, M.; Fernández-Simón, E.; Piñol-Jurado, P.; Badimon, L.; Wollin, L.; Lleixà, C.; Gallardo, E.; Olivé, M.; et al. Nintedanib Reduces Muscle Fibrosis and Improves Muscle Function of the Alpha-Sarcoglycan-Deficient Mice. Biomedicines 2022, 10, 2629. [Google Scholar] [CrossRef]
- Croce, S.; Cobianchi, L.; Zoro, T.; Mas, F.D.; Cornaglia, A.I.; Lenta, E.; Acquafredda, G.; De Silvestri, A.; Avanzini, M.A.; Visai, L.; et al. Mesenchymal Stromal Cell on Liver Decellularised Extracellular Matrix for Tissue Engineering. Biomedicines 2022, 10, 2817. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, M.; Turnbull, T.; Sebastian, S.; Kempson, I. The MTT Assay: Utility, Limitations, Pitfalls, and Interpretation in Bulk and Single-Cell Analysis. Int. J. Mol. Sci. 2021, 22, 12827. [Google Scholar] [CrossRef]
- Chen, Q.; Zhang, M.; Li, Y.; Xu, D.; Wang, Y.; Song, A.; Zhu, B.; Huang, Y.; Zheng, J.C. CXCR7 Mediates Neural Progenitor Cells Migration to CXCL12 Independent of CXCR4. Stem Cells 2015, 33, 2574–2585. [Google Scholar] [CrossRef]
- Zanella, E.R.; Grassi, E.; Trusolino, L. Towards precision oncology with patient-derived xenografts. Nat. Rev. Clin. Oncol. 2022, 19, 719–732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Yan, Y.; Wu, Y.; Yang, H.; Zhu, P.; Yan, F.; Zhao, R.; Tian, P.; Wang, T.; Fan, Q.; et al. Medicinal herbs for the treatment of anxiety: A systematic review and network meta-analysis. Pharmacol. Res. 2022, 179, 106204. [Google Scholar] [CrossRef] [PubMed]
- Scher, J.U.; Ogdie, A.; Merola, J.F.; Ritchlin, C. Preventing psoriatic arthritis: Focusing on patients with psoriasis at increased risk of transition. Nat. Rev. Rheumatol. 2019, 15, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate cancer. Lancet 2021, 398, 1075–1090. [Google Scholar] [CrossRef]
- Lu, X.; Horner, J.W.; Paul, E.; Shang, X.; Troncoso, P.; Deng, P.; Jiang, S.; Chang, Q.; Spring, D.J.; Sharma, P.; et al. Effective combinatorial immunotherapy for castration-resistant prostate cancer. Nature 2017, 543, 728–732, Corrected in Nature 2017, 545, 116. [Google Scholar] [CrossRef] [PubMed]
- Pala, L.; De Pas, T.; Conforti, F. Boosting anticancer immunotherapy through androgen receptor blockade. Cancer Cell 2022, 40, 455–457. [Google Scholar] [CrossRef]
- Becherer, U.; Medart, M.R.; Schirra, C.; Krause, E.; Stevens, D.; Rettig, J. Regulated exocytosis in chromaffin cells and cytotoxic T lymphocytes: How similar are they? Cell Calcium. 2012, 52, 303–312. [Google Scholar] [CrossRef]
- Skurikhin, E.G.; Pershina, O.; Ermakova, N.; Pakhomova, A.; Widera, D.; Zhukova, M.; Pan, E.; Sandrikina, L.; Kogai, L.; Kushlinskii, N.; et al. Reprogrammed CD8+ T-Lymphocytes Isolated from Bone Marrow Have Anticancer Potential in Lung Cancer. Biomedicines 2022, 10, 1450. [Google Scholar] [CrossRef]
- Baran, K.; Waśko, J.; Kryczka, J.; Boncela, J.; Jabłoński, S.; Kolesińska, B.; Brzeziańska-Lasota, E.; Kordiak, J. The Comparison of Serum Exosome Protein Profile in Diagnosis of NSCLC Patients. Int. J. Mol. Sci. 2023, 24, 13669. [Google Scholar] [CrossRef]
- Wang, G.C.; Gan, X.; Zeng, Y.Q.; Chen, X.; Kang, H.; Huang, S.-W.; Hu, W.-H. The Role of NCS1 in Immunotherapy and Prognosis of Human Cancer. Biomedicines 2023, 11, 2765. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the Roots of Cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef]
- Wang, L.; Luo, J.Y.; Li, B.; Tian, X.Y.; Chen, L.-J.; Huang, Y.; Liu, J.; Deng, D.; Lau, C.W.; Wan, S.; et al. Integrin-YAP/TAZ-JNK cascade mediates atheroprotective effect of unidirectional shear flow. Nature 2016, 540, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Haak, A.J.; Kostallari, E.; Sicard, D.; Ligresti, G.; Choi, K.M.; Caporarello, N.; Jones, D.L.; Tan, Q.; Meridew, J.; Espinosa, A.M.D.; et al. Selective YAP/TAZ inhibition in fibroblasts via dopamine receptor D1 agonism reverses fibrosis. Sci. Transl. Med. 2019, 11, eaau6296. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Xu, D.; Wang, S.; Wong, C.K.; Martinez-Fundichely, A.; Lee, C.J.; Cohen, S.; Park, J.; Hill, C.E.; Eng, K.; et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 2022, 376, eabe1505. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, A.; Walker, S.; Smith, J.; Patterson, K.; Dutt, A.; Ng, Y.M.; Thomas, H.D.; Wilson, L.; McCullough, B.; Jones, D.; et al. IKBKE activity enhances AR levels in advanced prostate cancer via modulation of the Hippo pathway. Nucleic Acids Res. 2020, 48, 5366–5382. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.R.; Thorburn, A. Autophagy and organelle homeostasis in cancer. Dev. Cell. 2021, 56, 906–918. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.R.; Jiang, Z.B.; Xu, C.; Meng, W.-Y.; Liu, P.; Zhang, Y.-Z.; Xie, C.; Xu, J.-Y.; Xie, Y.-J.; Liang, T.-L.; et al. Andrographolide suppresses non-small-cell lung cancer progression through induction of autophagy and antitumor immune response. Pharmacol. Res. 2022, 179, 106198. [Google Scholar] [CrossRef] [PubMed]
- Marzagalli, M.; Ebelt, N.D.; Manuel, E.R. Unraveling the crosstalk between melanoma and immune cells in the tumor microenvironment. Semin. Cancer Biol. 2019, 59, 236–250. [Google Scholar] [CrossRef] [PubMed]
- Deepak, K.; Vempati, R.; Nagaraju, G.P.; Dasari, V.R.; Nagini, S.; Rao, D.; Malla, R.R. Tumor microenvironment: Challenges and opportunities in targeting metastasis of triple negative breast cancer. Pharmacol. Res. 2020, 153, 104683. [Google Scholar] [CrossRef]
- Chen, Y.; Liu, B.; Wei, Y.; Kuang, D.M. Influence of gut and intratumoral microbiota on the immune microenvironment and anti-cancer therapy. Pharmacol. Res. 2021, 174, 105966. [Google Scholar] [CrossRef] [PubMed]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Wu, N.; Li, J.; Zhou, D.; Liang, J.; Cao, Q.; Guan, Z.; Xu, Y.; Jiang, N. YAP1 Regulates the YAP1/AR/PSA Axis through Autophagy in Castration-Resistant Prostate Cancer and Mediates T-Cell Immune and Inflammatory Cytokine Infiltration. Biomedicines 2024, 12, 661. https://doi.org/10.3390/biomedicines12030661
Wang Y, Wu N, Li J, Zhou D, Liang J, Cao Q, Guan Z, Xu Y, Jiang N. YAP1 Regulates the YAP1/AR/PSA Axis through Autophagy in Castration-Resistant Prostate Cancer and Mediates T-Cell Immune and Inflammatory Cytokine Infiltration. Biomedicines. 2024; 12(3):661. https://doi.org/10.3390/biomedicines12030661
Chicago/Turabian StyleWang, Youzhi, Ning Wu, Junbo Li, Diansheng Zhou, Jiaming Liang, Qian Cao, Zhaokai Guan, Yangyang Xu, and Ning Jiang. 2024. "YAP1 Regulates the YAP1/AR/PSA Axis through Autophagy in Castration-Resistant Prostate Cancer and Mediates T-Cell Immune and Inflammatory Cytokine Infiltration" Biomedicines 12, no. 3: 661. https://doi.org/10.3390/biomedicines12030661
APA StyleWang, Y., Wu, N., Li, J., Zhou, D., Liang, J., Cao, Q., Guan, Z., Xu, Y., & Jiang, N. (2024). YAP1 Regulates the YAP1/AR/PSA Axis through Autophagy in Castration-Resistant Prostate Cancer and Mediates T-Cell Immune and Inflammatory Cytokine Infiltration. Biomedicines, 12(3), 661. https://doi.org/10.3390/biomedicines12030661