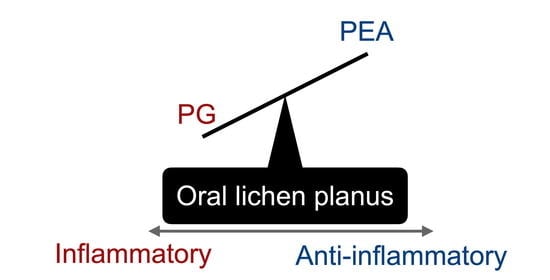
Relative Deficiency of Anti-Inflammatory N-Acylethanolamines Compared to Prostaglandins in Oral Lichen Planus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Biopsy Samples
2.2. RNA Extraction
2.3. Treatment of CAL27 and SCC-25 Human Squamous Carcinoma Cells with TNF-α, IL-8 and PEA
2.4. Real-Time qPCR (RT-qPCR)
2.5. Analysis of Oxylipins and N-Acylethanolamine (NAE) Derivatives
2.6. Statistics
3. Results
3.1. mRNA Levels of NAPE, PTGS2, FAAH and NAAA in Control and OLP Biopsy Samples
3.2. NAE Levels in Control and OLP Biopsy Samples
3.3. Oxylipin Levels in Control and OLP Biopsy Samples
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sugerman, P.B.; Savage, N.W.; Walsh, L.J.; Zhao, Z.Z.; Zhou, X.J.; Khan, A.; Seymour, G.J.; Bigby, M. The pathogenesis of oral lichen planus. Crit. Rev. Oral Biol. Med. 2002, 13, 350–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.; Tang, X.; Zheng, X.; Ge, S.; Wen, H.; Lin, X.; Chen, Z.; Lu, L. Global prevalence and incidence estimates of oral lichen planus: A systematic review and meta-analysis. JAMA Dermatol. 2020, 156, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Humberto, J.S.M.; Pavanin, J.V.; Rocha, M.; Motta, A.C.F. Cytokines, cortisol, and nitric oxide as salivary biomarkers in oral lichen planus: A systematic review. Braz. Oral Res. 2018, 32, e82. [Google Scholar] [CrossRef] [PubMed]
- Lysitsa, S.; Samson, J.; Gerber-Wicht, C.; Lang, U.; Lombardi, T. COX-2 expression in oral lichen planus. Dermatology 2008, 217, 150–155. [Google Scholar] [CrossRef]
- Abdel Hay, R.M.; Fawzy, M.M.; Metwally, D.; Kadry, D.; Ezzat, M.; Rashwan, W.; Rashed, L.A. DNA polymorphisms and tissue cyclooxygenase-2 expression in oral lichen planus: A case-control study. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 1122–1126. [Google Scholar] [CrossRef]
- Danielsson, K.; Ebrahimi, M.; Wahlin, Y.B.; Nylander, K.; Boldrup, L. Increased levels of COX-2 in oral lichen planus supports an autoimmune cause of the disease. J. Eur. Acad. Dermatol. Venereol. 2012, 26, 1415–1419. [Google Scholar] [CrossRef] [PubMed]
- Chankong, T.; Chotjumlong, P.; Sastraruji, T.; Pongsiriwet, S.; Iamaroon, A.; Krisanaprakornkit, S. Increased cyclooxygenase 2 expression in association with oral lichen planus severity. J. Dent. Sci. 2016, 11, 238–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ueda, N.; Tsuboi, K.; Uyama, T. Metabolism of endocannabinoids and related N-acylethanolamines: Canonical and alternative pathways. FEBS J. 2013, 280, 1874–1894. [Google Scholar] [CrossRef]
- Hansen, H.S. Effect of diet on tissue levels of palmitoylethanolamide. CNS Neurol. Disord. Drug. Targets 2013, 12, 17–25. [Google Scholar] [CrossRef]
- Devane, W.A.; Hanus, L.; Breuer, A.; Pertwee, R.G.; Stevenson, L.A.; Griffin, G.; Gibson, D.; Mandelbaum, A.; Etinger, A.; Mechoulam, R. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science 1992, 258, 1946–1949. [Google Scholar] [CrossRef]
- de Fonseca, F.R.; Navarro, M.; Gómez, R.; Escuredo, L.; Nava, F.; Fu, J.; Murillo-Rodríguez, E.; Giuffrida, A.; LoVerme, J.; Gaetani, S.; et al. An anorexic lipid mediator regulated by feeding. Nature 2001, 414, 209–212. [Google Scholar] [CrossRef] [Green Version]
- Lo Verme, J.; Fu, J.; Astarita, G.; La Rana, G.; Russo, R.; Calignano, A.; Piomelli, D. The nuclear receptor peroxisome proliferator-activated receptor-α mediates the anti-inflammatory actions of palmitoylethanolamide. Mol. Pharmacol. 2005, 67, 15–19. [Google Scholar] [CrossRef]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular actions of PPARα in lipid metabolism and inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costa, B.; Conti, S.; Giagnoni, G.; Colleoni, M. Therapeutic effect of the endogenous fatty acid amide, palmitoylethanolamide, in rat acute inflammation: Inhibition of nitric oxide and cyclo-oxygenase systems. Br. J. Pharmacol. 2002, 137, 413–420. [Google Scholar] [CrossRef]
- Gabrielsson, L.; Gouveia-Figueira, S.; Häggström, J.; Alhouayek, M.; Fowler, C.J. The anti-inflammatory compound palmitoylethanolamide inhibits prostaglandin and hydroxyeicosatetraenoic acid production by a macrophage cell line. Pharmacol. Res. Perspect. 2017, 5, e00300. [Google Scholar] [CrossRef]
- Aloe, L.; Leon, A.; Levi-Montalcini, R. A proposed autacoid mechanism controlling mastocyte behaviour. Agents Actions 1993, 39, C145–C147. [Google Scholar] [CrossRef] [PubMed]
- Gabrielsson, L.; Mattsson, S.; Fowler, C.J. Palmitoylethanolamide for the treatment of pain: Pharmacokinetics, safety and efficacy. Br. J. Clin. Pharmacol. 2016, 82, 932–942. [Google Scholar] [CrossRef] [Green Version]
- Eberlein, B.; Eicke, C.; Reinhardt, H.W.; Ring, J. Adjuvant treatment of atopic eczema: Assessment of an emollient containing N-palmitoylethanolamine (ATOPA study). J. Eur. Acad. Dermatol. Venereol. 2008, 22, 73–82. [Google Scholar] [CrossRef]
- Holt, S.; Comelli, F.; Costa, B.; Fowler, C.J. Inhibitors of fatty acid amide hydrolase reduce carrageenan-induced hind paw inflammation in pentobarbital-treated mice: Comparison with indomethacin and possible involvement of cannabinoid receptors. Br. J. Pharmacol. 2005, 146, 467–476. [Google Scholar] [CrossRef] [Green Version]
- Solorzano, C.; Zhu, C.; Battista, N.; Astarita, G.; Lodola, A.; Rivara, S.; Mor, M.; Russo, R.; Maccarrone, M.; Antonietti, F.; et al. Selective N-acylethanolamine-hydrolyzing acid amidase inhibition reveals a key role for endogenous palmitoylethanolamide in inflammation. Proc. Natl. Acad. Sci. USA 2009, 106, 20966–20971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, D.; Pearson, R.G.; Kurian, N.; Latif, M.L.; Garle, M.J.; Barrett, D.A.; Kendall, D.A.; Scammell, B.E.; Reeve, A.J.; Chapman, V. Characterisation of the cannabinoid receptor system in synovial tissue and fluid in patients with osteoarthritis and rheumatoid arthritis. Arthritis Res. Ther. 2008, 10, R43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suárez, J.; Romero-Zerbo, Y.; Márquez, L.; Rivera, P.; Iglesias, M.; Bermúdez-Silva, F.J.; Andreu, M.; de Fonseca, F.R. Ulcerative colitis impairs the acylethanolamide-based anti-inflammatory system reversal by 5-aminosalicylic acid and glucocorticoids. PLoS ONE 2012, 7, e37729. [Google Scholar] [CrossRef] [PubMed]
- Barry, A.; O’Halloran, K.D.; McKenna, J.P.; McCreary, C.; Harhen, B.; Kerr, D.M.; Finn, D.P.; Downer, E.J. Plasma N-acylethanolamine and endocannabinoid levels in burning mouth syndrome: Potential role in disease pathogenesis. J. Oral Pathol. Med. 2018, 47, 440–442. [Google Scholar] [CrossRef]
- Danielsson, K.; Coates, P.J.; Ebrahimi, M.; Nylander, E.; Wahlin, Y.B.; Nylander, K. Genes involved in epithelial differentiation and development are differentially expressed in oral and genital lichen planus epithelium compared to normal epithelium. Acta Derm. Venereol. 2014, 94, 526–530. [Google Scholar] [CrossRef] [Green Version]
- Gouveia-Figueira, S.; Danielsson, K.; Fowler, C.J. Changes in proportions of linoleic acid-derived oxylipins in oral lichen planus. Acta Derm. Venereol. 2019, 99, 1051–1052. [Google Scholar] [CrossRef]
- Gouveia-Figueira, S.; Nording, M.L. Validation of a tandem mass spectrometry method using combined extraction of 37 oxylipins and 14 endocannabinoid-related compounds including prostamides from biological matrices. Prostaglandins Other Lipid Mediat. 2015, 121, 110–121. [Google Scholar] [CrossRef]
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.r-project.org (accessed on 31 August 2020).
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Stat. Soc. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Karlsson, J.; Gouveia-Figueira, S.; Alhouayek, M.; Fowler, C.J. Effects of tumour necrosis factor α upon the metabolism of the endocannabinoid anandamide in prostate cancer cells. PLoS ONE 2017, 12, e0185011. [Google Scholar] [CrossRef] [Green Version]
- Aitchison, J. The statistical analysis of compositional data. J. R. Stat. Soc. B 1982, 44, 139–177. [Google Scholar] [CrossRef]
- Huang, S.M.; Bisogno, T.; Petros, T.J.; Chang, S.Y.; Zavitsanos, P.A.; Zipkin, R.E.; Sivakumar, R.; Coop, A.; Maeda, D.Y.; De Petrocellis, L.; et al. Identification of a new class of molecules, the arachidonyl amino acids, and characterization of one member that inhibits pain. J. Biol. Chem. 2001, 276, 42639–42644. [Google Scholar] [CrossRef] [Green Version]
- Capasso, R.; Izzo, A.; Fezza, F.; Pinto, A.; Capasso, F.; Mascolo, N.; Di Marzo, V. Inhibitory effect of palmitoylethanolamide on gastrointestinal motility in mice. Br. J. Pharmacol. 2001, 134, 945–950. [Google Scholar] [CrossRef] [Green Version]
- Baker, D.; Pryce, G.; Croxford, J.L.; Brown, P.; Pertwee, R.G.; Makriyannis, A.; Khanolkar, A.; Layward, L.; Fezza, F.; Bisogno, T.; et al. Endocannabinoids control spasticity in a multiple sclerosis model. FASEB J. 2001, 15, 300–302. [Google Scholar] [CrossRef] [Green Version]
- Rothman, K.J. No adjustments are needed for multiple comparisons. Epidemiology 1990, 1, 43–46. [Google Scholar] [CrossRef] [Green Version]
- Bonsale, R.; Seyed Sharifi, R.; Dirandeh, E.; Hedayat, N.; Mojtahedin, A.; Ghorbanalinia, M.; Abolghasemi, A. Endocannabinoids as endometrial inflammatory markers in lactating Holstein cows. Reprod. Domest. Anim. 2018, 53, 769–775. [Google Scholar] [CrossRef] [PubMed]
- Santarelli, A.; Mascitti, M.; Rubini, C.; Bambini, F.; Zizzi, A.; Offidani, A.; Ganzetti, G.; Laino, L.; Cicciù, M.; Lo Muzio, L. Active inflammatory biomarkers in oral lichen planus. Int. J. Immunopathol. Pharmacol. 2015, 28, 562–568. [Google Scholar] [CrossRef] [Green Version]
- Singh, P.; Grover, J.; Byatnal, A.A.; Guddattu, V.; Radhakrishnan, R.; Solomon, M.C. Elucidating the role of cyclooxygenase-2 in the pathogenesis of oral lichen planus—An immunohistochemical study with supportive histochemical analysis. J. Oral Pathol. Med. 2017, 46, 381–386. [Google Scholar] [CrossRef] [PubMed]
- Subbarayan, V.; Sabichi, A.L.; Llansa, N.; Lippman, S.M.; Menter, D.G. Differential expression of cyclooxygenase-2 and its regulation by tumor necrosis factor-α in normal and malignant prostate cells. Cancer Res. 2001, 61, 2720–2726. [Google Scholar]
- Alhouayek, M.; Boldrup, L.; Fowler, C.J. Altered mRNA expression of genes involved in endocannabinoid signalling in squamous cell carcinoma of the oral tongue. Cancer Investig. 2019, 37, 327–338. [Google Scholar] [CrossRef]
- Henneicke-von Zepelin, H.H.; Schröder, J.M.; Smíd, P.; Reusch, M.K.; Christophers, E. Metabolism of arachidonic acid by human epidermal cells depends upon maturational stage. J. Investig. Dermatol. 1991, 97, 291–297. [Google Scholar] [CrossRef] [Green Version]
- Johnson, G.K.; Payne, J.B.; Fili, J.M.; Reinhardt, R.A.; Organ, C.C.; Slager, S.L. Development of smokeless tobacco-induced oral mucosal lesions. J. Oral Pathol. Med. 1998, 27, 388–394. [Google Scholar] [CrossRef]
- Annuzzi, G.; Piscitelli, F.; Di Marino, L.; Patti, L.; Giacco, R.; Costabile, G.; Bozzetto, L.; Riccardi, G.; Verde, R.; Petrosino, S.; et al. Differential alterations of the concentrations of endocannabinoids and related lipids in the subcutaneous adipose tissue of obese diabetic patients. Lipids Health Dis. 2010, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Stensson, N.; Ghafouri, B.; Ghafouri, N.; Gerdle, B. High levels of endogenous lipid mediators (N-acylethanolamines) in women with chronic widespread pain during acute tissue trauma. Mol. Pain 2016, 12. [Google Scholar] [CrossRef] [Green Version]
- Rankin, L.; Fowler, C.J. The basal pharmacology of palmitoylethanolamide. Int. J. Mol. Sci. 2020, 21, 7942. [Google Scholar] [CrossRef]
- Guida, G.; De Martino, M.; De Fabiani, A.; Cantieri, L.A.; Alexandre, A.; Vassallo, G.M.; Rogai, M.; Lanaia, F.; Petrosino, S. La palmitoilethanolamida (Normast) en el dolor neuropático crónico por lumbociatalgia de tipo compresivo: Estudio clínico multicéntrico. Dolor 2010, 25, 35–42. [Google Scholar]
- Steels, E.; Venkatesh, R.; Steels, E.; Vitetta, G.; Vitetta, L. A double-blind randomized placebo controlled study assessing safety, tolerability and efficacy of palmitoylethanolamide for symptoms of knee osteoarthritis. Inflammopharmacology 2019, 27, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Ottaviani, G.; Rupel, K.; Gobbo, M.; Poropat, A.; Zoi, V.; Faraon, M.; Di Lenarda, R.; Biasotto, M. Efficacy of ultramicronized palmitoylethanolamide in burning mouth syndrome-affected patients: A preliminary randomized double-blind controlled trial. Clin. Oral Investig. 2019, 23, 2743–2750. [Google Scholar] [CrossRef]
- Orefice, N.S.; Alhouayek, M.; Carotenuto, A.; Montella, S.; Barbato, F.; Comelli, A.; Calignano, A.; Muccioli, G.G.; Orefice, G. Oral palmitoylethanolamide treatment is associated with reduced cutaneous adverse effects of interferon-β1a and circulating proinflammatory cytokines in relapsing-remitting multiple sclerosis. Neurotherapeutics 2016, 13, 428–438. [Google Scholar] [CrossRef]





| Controls | OLP Patients | ||||
|---|---|---|---|---|---|
| Gender | Age | Gender | Age | OLP Type | Localization |
| M | 69 | M | 67 | reticular, atrophic, plaque, ulcerous | oral, genital |
| F | 69 | F | 66 | reticular, atrophic | oral, genital |
| M | 49 | F | 63 | reticular, atrophic, plaque | oral, genital |
| F | 53 | M | 61 | reticular, atrophic | oral, skin |
| F | 50 | M | 58 | reticular | oral |
| F | 47 | F a | 81 | reticular, atrophic | oral |
| F a | 67 | M | 45 | reticular, atrophic | oral, genital |
| F | 45 | F | 63 | reticular, atrophic | oral |
| F | 44 | F | 73 | reticular, atrophic | oral, genital |
| F | 39 | F | 74 | reticular, atrophic | oral, genital |
| M | 66 | F | 64 | reticular, atrophic | oral, genital |
| F | 64 | F | 64 | reticular, atrophic | oral, skin |
| F | 61 | M | 67 | reticular | oral |
| F | 73 | F b | 44 | reticular | oral, genital |
| F | 68 | F | 68 | reticular, atrophic, ulcerous | oral |
| Gene (Product). | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Efficiency |
|---|---|---|---|
| RPL19 (Ribosomal protein 19) | CAC ATC CAC AAG CTG AAG GCA | CTT GCG TGC TTC CTT GGT CT | 99% |
| NAPEPLD (NAPE-PLD) | ACT GGT TAT TGC CCT GCT TT | AAT CCT TAC AGC TTC TTC TGG G | 99% |
| PTGS2 (COX-2) | AGC AGG CAG ATG AAA TAC CAG | ACC AGA AGG GCA GGA TAC A | 93% |
| NAAA (NAAA) | ATG GAG CGT GGT TCC GAG TT | AGG CTG AGG TTT GCT TGT CCT | 99% |
| FAAH (FAAH) | CAC ACG CTG GTT CCC TTC TT | GGG TCC ACG AAA TCA CCT TTG A | 99% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rankin, L.; Gouveia-Figueira, S.; Danielsson, K.P.; Fowler, C.J. Relative Deficiency of Anti-Inflammatory N-Acylethanolamines Compared to Prostaglandins in Oral Lichen Planus. Biomedicines 2020, 8, 481. https://doi.org/10.3390/biomedicines8110481
Rankin L, Gouveia-Figueira S, Danielsson KP, Fowler CJ. Relative Deficiency of Anti-Inflammatory N-Acylethanolamines Compared to Prostaglandins in Oral Lichen Planus. Biomedicines. 2020; 8(11):481. https://doi.org/10.3390/biomedicines8110481
Chicago/Turabian StyleRankin, Linda, Sandra Gouveia-Figueira, Karin P. Danielsson, and Christopher J. Fowler. 2020. "Relative Deficiency of Anti-Inflammatory N-Acylethanolamines Compared to Prostaglandins in Oral Lichen Planus" Biomedicines 8, no. 11: 481. https://doi.org/10.3390/biomedicines8110481
APA StyleRankin, L., Gouveia-Figueira, S., Danielsson, K. P., & Fowler, C. J. (2020). Relative Deficiency of Anti-Inflammatory N-Acylethanolamines Compared to Prostaglandins in Oral Lichen Planus. Biomedicines, 8(11), 481. https://doi.org/10.3390/biomedicines8110481