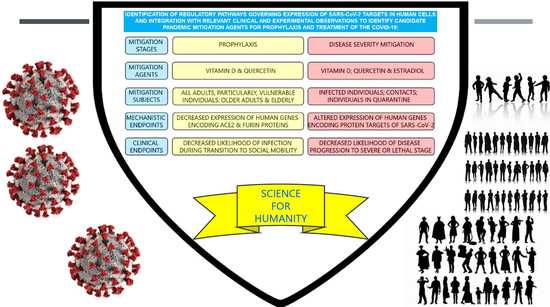
Tripartite Combination of Candidate Pandemic Mitigation Agents: Vitamin D, Quercetin, and Estradiol Manifest Properties of Medicinal Agents for Targeted Mitigation of the COVID-19 Pandemic Defined by Genomics-Guided Tracing of SARS-CoV-2 Targets in Human Cells
Abstract
:1. Introduction
2. Methods
2.1. Data Source and Analytical Protocols
2.2. Statistical Analyses of the Publicly Available Datasets
3. Results and Discussion
3.1. Enrichr-Guided Gene Set Enrichment Analyses (GSEA) of Genomic Features Associated with the ACE2 and FURIN Genes
3.2. Identifications of the Enriched Records of Transcription Factor-Binding Sites Affecting the ACE2 and FURIN Expression
3.3. Identification of the VDR and HIF1a Genes as Putative Repressors of the ACE2 Expression
3.4. GSEA Identify Estradiol and Quercetin as Putative Candidate Coronavirus Infection Mitigation Agents
3.5. Confirmation of the Estradiol and Quercetin Activities as Potential Candidate Coronavirus Infection Mitigation Agents
3.6. Inference of Potential Interference of the Quercetin, Vitamin D, and Estradiol with Functions of SARS-CoV-2 Proteins in Human Cells
3.7. Potential Mechanisms Affecting Gene Expression Inferred from Transgenic Mouse Models and Observed in Pathophysiologically and Therapeutically Relevant Mouse and Human Cells
3.8. Is Vitamin D Deficiency a Potential Risk Factor for Increased Disease Severity in Older Adults and Elderly Individuals?
3.9. Clinical and Experimental Observations Appear Highly Compatible with the Results of Genomic-Guided Mapping of COVID-19 Targets in Human Cells
3.10. Limitations
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef] [PubMed]
- Walls, A.C.; Park, Y.J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Shang, J.; Ye, G.; Shi, K.; Wan, Y.; Luo, C.; Aihara, H.; Geng, Q.; Auerbach, A.; Li, F. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448. [Google Scholar] [CrossRef] [Green Version]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef] [Green Version]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [Green Version]
- Glinsky, G.V. Human-specific features of pluripotency regulatory networks link NANOG with fetal and adult brain development. BioRxiv 2017. [Google Scholar] [CrossRef] [Green Version]
- Glinsky, G.V. Contribution of transposable elements and distal enhancers to evolution of human-specific features of interphase chromatin architecture in embryonic stem cells. Chromosome Res. 2018, 26, 61–84. [Google Scholar] [CrossRef]
- Glinsky, G.; Durruthy-Durruthy, J.; Wossidlo, M.; Grow, E.J.; Weirather, J.L.; Au, K.F.; Wysocka, J.; Sebastiano, V. Single cell expression analysis of primate-specific retroviruses-derived HPAT lincRNAs in viable human blastocysts identifies embryonic cells co-expressing genetic markers of multiple lineages. Heliyon 2018, 4, e00667. [Google Scholar] [CrossRef]
- Glinsky, G.V.; Barakat, T.S. The evolution of Great Apes has shaped the functional enhancers’ landscape in human embryonic stem cells. Stem Cell Res. 2019, 37, 101456. [Google Scholar] [CrossRef] [PubMed]
- Glinsky, G.V. A catalogue of 59,732 human-specific regulatory sequences reveals unique to human regulatory patterns associated with virus-interacting proteins, pluripotency and brain development. DNA Cell Biol. 2020, 39, 126–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glinsky, G.V. Impacts of genomic networks governed by human-specific regulatory sequences and genetic loci harboring fixed human-specific neuro-regulatory single nucleotide mutations on phenotypic traits of Modern Humans. bioRxiv 2020, 848762. [Google Scholar] [CrossRef] [Green Version]
- Guffanti, G.; Bartlett, A.; Klengel, T.; Klengel, C.; Hunter, R.; Glinsky, G.; Macciardi, F. Novel bioinformatics approach identifies transcriptional profiles of lineage-specific transposable elements at distinct loci in the human dorsolateral prefrontal cortex. Mol. Biol. Evol. 2018, 35, 2435–2453. [Google Scholar] [CrossRef] [PubMed]
- Tavazoie, S.; Hughes, J.D.; Campbell, M.J.; Cho, R.J.; Church, G.M. Systematic determination of genetic network architecture. Nat. Genet. 1999, 22, 281–285. [Google Scholar] [CrossRef]
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nature Communications 9. 2018, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Reghunathan, R.; Jayapal, M.; Hsu, L.Y.; Chng, H.H.; Tai, D.; Leung, B.P.; Melendez, A.J. Expression profile of immune response genes in patients with Severe Acute Respiratory Syndrome. BMC Immunol. 2005, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Helming, L.; B#xF6;se, J.; Ehrchen, J.; Schiebe, S.; Frahm, T.; Geffers, R.; Probst-Kepper, M.; Balling, R.; Lengeling, A. 1alpha,25-Dihydroxyvitamin D3 is a potent suppressor of interferon gamma-mediated macrophage activation. Blood 2005, 106, 4351–4358. [Google Scholar] [CrossRef] [Green Version]
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; O’meara, M.J.; Guo, J.Z.; Swaney, D.L.; Tummino, T.A.; Huttenhain, R.; et al. A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature 2020, 1–13. [Google Scholar] [CrossRef]
- Seuter, S.; Neme, A.; Carlberg, C. Epigenome-wide effects of vitamin D and their impact on the transcriptome of human monocytes involve CTCF. Nucleic Acids Res. 2016, 44, 4090–4104. [Google Scholar] [CrossRef] [Green Version]
- Neme, A.; Seuter, S.; Carlberg, C. Selective regulation of biological processes by vitamin D based on the spatio-temporal cistrome of its receptor. Biochim. Biophys. Acta 2017, 1860, 952–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennel, K.A.; Drake, M.T.; Hurley, D.L. Vitamin D deficiency in adults: When to test and how to treat. Mayo Clin Proc. 2010, 85, 752–757. [Google Scholar] [CrossRef] [Green Version]
- Ginde, A.A.; Liu, M.C.; Camargo, C.A., Jr. Demographic differences and trends of vitamin D insufficiency in the US population, 1988-2004. Arch. Intern. Med. 2009, 169, 626–632. [Google Scholar] [CrossRef] [Green Version]
- Lips, P. Vitamin D deficiency and secondary hyperparathyroidism in the elderly: Consequences for bone loss and fractures and therapeutic implications. Endocr Rev. 2001, 22, 477–501. [Google Scholar] [CrossRef] [PubMed]
- Autier, P.; Gandini, S. Vitamin D supplementation and total mortality: A meta-analysis of randomized controlled trials. Arch. Intern. Med. 2017, 167, 1730–1737. [Google Scholar] [CrossRef] [PubMed]
- Ginde, A.A.; Scragg, R.; Schwartz, R.S.; Camargo, C.A., Jr. Prospective study of serum 25-hydroxyvitamin d level, cardiovascular disease mortality, and all-cause mortality in older U.S. Adults. J. Am. Geriatr. Soc. 2009, 57, 1595–1603. [Google Scholar] [CrossRef]
- Heath, A.K.; Kim, I.Y.; Hodge, A.M.; English, D.R.; Muller, D.C. Vitamin D status and mortality: A systematic review of observational studies. Int. J. Environ. Res. Public Health 2019, 16, 383. [Google Scholar] [CrossRef] [Green Version]
- Caristia, S.; Filigheddu, N.; Barone-Adesi, F.; Sarro, A.; Testa, T.; Magnani, C.; Aimaretti, G.; Faggiano, F.; Marzullo, P. Vitamin D as a biomarker of ill health among the over-50s: A systematic review of cohort studies. Nutrients 2019, 11, E2384. [Google Scholar] [CrossRef] [Green Version]
- Grant, W.B.; Lahore, H.; McDonnell, S.L.; Baggerly, C.A.; French, C.B.; Aliano, J.L.; Bhattoa, H.P. Evidence that Vitamin D Supplementation Could Reduce Risk of Influenza and COVID-19 Infections and Deaths. Nutrients 2020, 12, 988. [Google Scholar] [CrossRef] [Green Version]
- Fabbri, A.; Infante, M.; Ricordi, C. Editorial―Vitamin D status: A key modulator of innate immunity and natural defense from acute viral respiratory infections. Eur. Rev. Med Pharmacol. Sci. 2020, 24, 4048–4052. [Google Scholar]
- Calder, P.C.; Carr, A.C.; Gombart, A.F.; Eggersdorfer, M. Optimal nutritional status for a well-functioning immune system is an important factor to protect against viral infections. Nutrients 2020, 12, 1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daneshkhan, A.; Agrawal, V.; Eshein, A.; Subramanian, H.; Roy, H.K.; Backman, V. The possible role of Vitamin D in suppressing cytokine storm and associated mortality in COVID-19 patients. MedRxiv 2020. [Google Scholar] [CrossRef]
- Wambier, C.G.; Goren, A. SARS-COV-2 infection is likely to be androgen mediated. J. Am. Acad Dermatol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Fett, C.; Mack, M.; Ten Eyck, P.P.; Meyerholz, D.K.; Perlman, S. Sex-Based Differences in Susceptibility to Severe Acute Respiratory Syndrome Coronavirus Infection. J. Immunol. 2017, 198, 4046–4053. [Google Scholar] [CrossRef]
- Wang, X.; Dhindsa, R.; Povysil, G.; Zoghbi, A.; Motelow, J.; Hostyk, J.; Goldstein, D. Transcriptional inhibition of host viral entry proteins as a therapeutic strategy for SARS-CoV-2. Preprints 2020. [Google Scholar] [CrossRef]
- McMichael, T.M.; Currie, D.W.; Clark, S.; Pogosjans, S.; Kay, M.; Schwartz, N.G.; Lewis, J.; Baer, A.; Kawakami, V.; Lukoff, M.D.; et al. Epidemiology of Covid-19 in a Long-Term Care Facility in King County, Washington. N. Engl. J. Med. 2020, in press. [Google Scholar] [CrossRef]
- Richardson, S.; Hirsch, J.S.; Narasimhan, M.; Crawford, J.M.; McGinn, T.; Davidson, K.W.; Barnaby, D.P.; Becker, L.B.; Chelico, J.D.; Cohen, S.L.; et al. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized with COVID-19 in the New York City Area. JAMA 2020, in press. [Google Scholar] [CrossRef]
- Shi, S.; Qin, M.; Shen, B.; Cai, Y.; Liu, T.; Yang, F.; Gong, W.; Liu, X.; Liang, J.; Zhao, Q.; et al. Association of Cardiac Injury with Mortality in Hospitalized Patients with COVID-19 in Wuhan, China. JAMA Cardiol. 2020, in press. [Google Scholar] [CrossRef] [Green Version]
- Paniz-Mondolfi, A.; Bryce, C.; Grimes, Z.; Gordon, R.E.; Reidy, J.; Lednicky, J.; Sordillo, E.M.; Fowkes, M. Central Nervous System Involvement by Severe Acute Respiratory Syndrome Coronavirus -2 (SARS-CoV-2). J. Med. Virol. 2020. [Google Scholar] [CrossRef] [Green Version]
- Sardu, C.; Gambardella, J.; Morelli, M.B.; Wang, X.; Marfella, R.; Santulli, G. Is COVID-19 an Endothelial Disease? Clinical and Basic Evidence. Preprints 2020. [Google Scholar] [CrossRef]
- Tang, N.; Li, D.; Wang, X.; Sun, Z. Abnormal Coagulation Parameters are Associated with Poor Prognosis in Patients with Novel Coronavirus Pneumonia. J. Thromb. Haemost. 2020, 18, 844–847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.; Smith, J.C. Repurposing Therapeutics for COVID-19: Supercomputer-Based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface. ChemRxiv. 2020. [Google Scholar] [CrossRef]
- Yi, L.; Li, Z.; Yuan, K.; Qu, X.; Chen, J.; Wang, G.; Zhang, H.; Luo, H.; Zhu, L.; Jiang, P.; et al. Small molecules blocking the entry of severe acute respiratory syndrome coronavirus into host cells. J. Virol. 2004, 78, 11334–11339. [Google Scholar] [CrossRef] [Green Version]
- Brielle, E.S.; Schneidman-Duhovny, D.; Linial, M. The SARS-CoV-2 exerts a distinctive strategy for interacting with the ACE2 human receptor. bioRxiv Prepr. 2020. [Google Scholar] [CrossRef] [PubMed]
- Jean, F.; Stella, K.; Thomas, L.; Liu, G.; Xiang, Y.; Reason, A.J.; Thomas, G. Alpha1-Antitrypsin Portland, a bioengineered serpin highly selective for furin: Application as an antipathogenic agent. Proc. Natl. Acad. Sci. USA 1998, 95, 7293–7298. [Google Scholar] [CrossRef] [Green Version]














| Therapeutic Modality | Number of Affected Genes | Percent of All SARS-CoV-2 Targets * |
|---|---|---|
| Vitamin D (all affected genes) | 84 ** | 25.30 |
| Quercetin (all affected genes) | 98 ** | 29.52 |
| Estradiol (all affected genes) | 203 ** | 61.14 |
| Vitamin D and Estradiol | 32 | 9.64 |
| Vitamin D and Quercetin | 6 | 1.81 |
| Quercetin and Estradiol | 57 | 17.17 |
| Vitamin D/Quercetin/Estradiol | 22 | 6.63 |
| Estradiol only | 90 | 27.11 |
| Quercetin only | 13 | 3.92 |
| Vitamin D only | 24 | 7.23 |
| All SARS-CoV-2 targets affected by Vitamin D/Quercetin/Estradiol | 244 ** | 73.49 |
| Therapeutic Modality | Number of Affected SARS-CoV-2 Proteins (33% Targets’ Threshold) | Percent of All SARS-CoV-2 Proteins (n = 27) | Number of Affected SARS-CoV-2 Proteins (50% Targets’ Threshold) | Percent of All SARS-CoV-2 Proteins (n = 27) | Number of Affected SARS-CoV-2 Proteins (70% Targets’ Threshold) | Percent of All SARS-CoV-2 Proteins (n = 27) |
|---|---|---|---|---|---|---|
| Vitamin D (all affected targets) | 10 | 37.04 | 4 | 14.81 | 1 | 3.70 |
| Quercetin (all affected targets) | 12 | 44.44 | 6 | 22.22 | 2 | 7.41 |
| Vitamin D and Quercetin | 23* | 85.19 | 11 | 40.74 | 3 | 11.11 |
| Estradiol (all affected targets) | 26 | 96.30 | 23 | 85.19 | 5 | 18.52 |
| Vitamin D/ Quercetin/Estradiol | 26 | 96.30 | 24 | 88.89 | 14 * | 51.85 |
| Testosterone | 8 | 29.63 | 4 | 14.81 | 1 | 3.70 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Glinsky, G.V. Tripartite Combination of Candidate Pandemic Mitigation Agents: Vitamin D, Quercetin, and Estradiol Manifest Properties of Medicinal Agents for Targeted Mitigation of the COVID-19 Pandemic Defined by Genomics-Guided Tracing of SARS-CoV-2 Targets in Human Cells. Biomedicines 2020, 8, 129. https://doi.org/10.3390/biomedicines8050129
Glinsky GV. Tripartite Combination of Candidate Pandemic Mitigation Agents: Vitamin D, Quercetin, and Estradiol Manifest Properties of Medicinal Agents for Targeted Mitigation of the COVID-19 Pandemic Defined by Genomics-Guided Tracing of SARS-CoV-2 Targets in Human Cells. Biomedicines. 2020; 8(5):129. https://doi.org/10.3390/biomedicines8050129
Chicago/Turabian StyleGlinsky, Gennadi V. 2020. "Tripartite Combination of Candidate Pandemic Mitigation Agents: Vitamin D, Quercetin, and Estradiol Manifest Properties of Medicinal Agents for Targeted Mitigation of the COVID-19 Pandemic Defined by Genomics-Guided Tracing of SARS-CoV-2 Targets in Human Cells" Biomedicines 8, no. 5: 129. https://doi.org/10.3390/biomedicines8050129
APA StyleGlinsky, G. V. (2020). Tripartite Combination of Candidate Pandemic Mitigation Agents: Vitamin D, Quercetin, and Estradiol Manifest Properties of Medicinal Agents for Targeted Mitigation of the COVID-19 Pandemic Defined by Genomics-Guided Tracing of SARS-CoV-2 Targets in Human Cells. Biomedicines, 8(5), 129. https://doi.org/10.3390/biomedicines8050129