Novel Biomimetic Human TLR2-Derived Peptides for Potential Targeting of Lipoteichoic Acid: An In Silico Assessment
Abstract
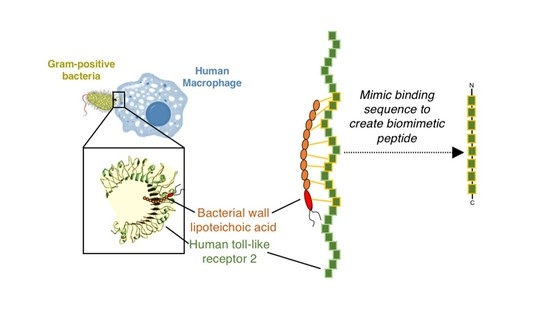
:1. Introduction
2. Materials and Methods
2.1. Receptor and Ligand Acquisition/Design and Preparation
2.2. Molecular Docking
2.3. Molecular Dynamics Simulation
2.4. Post-Dynamic Analyses
- Eele
- Electrostatic potential energy from Coulomb forces
- Egas
- Gas-phase energy (based on FF14SB force field terms)
- Eint
- Internal energy
- EvdW
- van der Waals energy
- Gsol
- Solvation free energy
- GGB
- Polar solvation energy
- GSA
- Non-polar solvation energy
- S
- Total entropy of solute
- SASA
- Solvent accessible surface area (water probe radius of 1.4 Å)
- T
- Total entropy of temperature
3. Results and Discussion
3.1. Binding Affinity of TLR2 to LTA
3.2. Molecular Dynamics of TLR2/LTA Complex for Acquisition of Binding Site Amino Acid Residues
3.3. Design of Potential Biomimetic TLR2-Derived Targeting Peptides
3.4. Binding Affinity of Biomimetic TLR2-Derived Peptides to LTA
3.5. Molecular Dynamics, Stability, Thermodynamics and Per-Residue Binding Free Energy of BTp /LTA Complexes
3.6. Lipophilicity of BTp2
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization. The Top 10 Causes of Death. Available online: https://www.who.int/news-room/fact-sheets/detail/the-top-10-causes-of-death (accessed on 14 December 2020).
- Lee, N.Y.; Ko, W.C.; Hsueh, P.R. Nanoparticles in the Treatment of Infections Caused by Multidrug-Resistant Organisms. Front. Pharmacol. 2019, 10, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Fair, R.J.; Tor, Y. Antibiotics and Bacterial Resistance in the 21st Century. Perspect. Medicin. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Andrea, M.M.; Fraziano, M.; Thaller, M.C.; Rossolini, G.M. The Urgent Need for Novel Antimicrobial Agents and Strategies to Fight Antibiotic Resistance. Antibiotics 2019, 8, 254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mine, Y.; Munir, H.; Nakanishi, Y.; Sugiyama, D. Biomimetic Peptides for the Treatment of Cancer. Anticancer Res. 2016, 36, 3565–3570. [Google Scholar] [PubMed]
- Jackman, J.A.; Yoon, B.K.; Ouyang, L.; Wang, N.; Ferhan, A.R.; Kim, J.; Majima, T.; Cho, N.J. Biomimetic Nanomaterial Strategies for Virus Targeting: Antiviral Therapies and Vaccines. Adv. Funct. Mater. 2020, 2008352. [Google Scholar] [CrossRef]
- Nanomik Biotechnology Protects Crops from Fungi Using Biomimetic Principles. Available online: https://www.raycandersonfoundation.org/articles/nanomik-biotechnology-protects-crops-from-fungi-using-biomimetic-principles (accessed on 15 December 2020).
- Chen, Y.X.; Wei, C.X.; Lyu, Y.Q.; Chen, H.Z.; Jiang, G.; Gao, X.L. Biomimetic Drug-Delivery Systems for the Management of Brain Diseases. Biomater. Sci. 2020, 8, 1073–1088. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Chen, S.; Zhang, J. Bioinspired and Biomimetic Nanotherapies for the Treatment of Infectious Diseases. Front. Pharmacol. 2019, 10, 751. [Google Scholar] [CrossRef]
- Li, A.; Zhao, J.; Fu, J.; Cai, J.; Zhang, P. Recent Advances of Biomimetic Nano-Systems in the Diagnosis and Treatment of Tumor. Asian J. Pharm. Sci. 2021, 16, 161–174. [Google Scholar] [CrossRef]
- Ibrahim, U.H.; Devnarain, N.; Govender, T. Biomimetic Strategies for Enhancing Synthesis and Delivery of Antibacterial Nanosystems. Int. J. Pharm. 2021, 596, 120276. [Google Scholar] [CrossRef]
- Ahmad, S.; Munir, S.; Zeb, N.; Ullah, A.; Khan, B.; Ali, J.; Bilal, M.; Omer, M.; Alamzeb, M.; Salman, S.M.; et al. Green Nanotechnology: A Review on Green Synthesis of Silver Nanoparticles—An Ecofriendly Approach. Int. J. Nanomed. 2019, 14, 5087–5107. [Google Scholar] [CrossRef] [Green Version]
- Saxena, J.; Sharma, P.; Singh, A. Biomimetic Synthesis of AgNPs from Penicillium Chrysogenum Strain FGCC/BLS1 by Optimising Physico-Cultural Conditions and Assessment of Their Antimicrobial Potential. IET Nanobiotechnol. 2017, 11, 576–583. [Google Scholar] [CrossRef]
- Li, L.L.; Xu, J.H.; Qi, G.B.; Zhao, X.; Yu, F.; Wang, H. Core-Shell Supramolecular Gelatin Nanoparticles for Adaptive and “on-Demand” Antibiotic Delivery. ACS Nano 2014, 8, 4975–4983. [Google Scholar] [CrossRef] [PubMed]
- Fulgione, A.; Ianniello, F.; Papaianni, M.; Contaldi, F.; Sgamma, T.; Giannini, C.; Pastore, S.; Velotta, R.; Ventura, B.D.; Roveri, N.; et al. Hydroxyapatite Nanocrystals Are an Active Carrier for Salmonella Bacteriophages. Int. J. Nanomed. 2019, 14, 2219–2232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Gao, W.; Chen, Y.; Escajadillo, T.; Ungerleider, J.; Fang, R.H.; Christman, K.; Nizet, V.; Zhang, L. Self-Assembled Colloidal Gel Using Cell Membrane-Coated Nanosponges as Building Blocks. ACS Nano 2017, 11, 11923–11930. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.G.; Shin, H.; Lim, D.W. Biomimetic Scaffolds for Tissue Engineering. Adv. Funct. Mater. 2012, 22, 2446–2468. [Google Scholar] [CrossRef]
- Qian, C.; Cao, X. Regulation of Toll-like Receptor Signaling Pathways in Innate Immune Responses. Ann. N. Y. Acad. Sci. 2013, 1283, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Javaid, N.; Yasmeen, F.; Choi, S. Toll-like Receptors and Relevant Emerging Therapeutics with Reference to Delivery Methods. Pharmaceutics 2019, 11, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, J.Y.; Nan, X.; Jin, M.S.; Youn, S.J.; Ryu, Y.H.; Mah, S.; Han, S.H.; Lee, H.; Paik, S.G.; Lee, J.O. Recognition of Lipopeptide Patterns by Toll-like Receptor 2-Toll-like Receptor 6 Heterodimer. Immunity 2009, 31, 873–884. [Google Scholar] [CrossRef] [Green Version]
- Goodsell, D.S.; Zardecki, C.; Di Costanzo, L.; Duarte, J.M.; Hudson, B.P.; Persikova, I.; Segura, J.; Shao, C.; Voigt, M.; Westbrook, J.D.; et al. RCSB Protein Data Bank: Enabling Biomedical Research and Drug Discovery. Protein Sci. 2020, 29, 52–65. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Thiessen, P.A.; Cheng, T.; Zhang, J.; Gindulyte, A.; Bolton, E.E. Pug-View: Programmatic Access to Chemical Annotations Integrated in PubChem. J. Cheminform. 2019, 11, 56. [Google Scholar] [CrossRef] [Green Version]
- Rayan, B.; Rayan, A. Avogadro Program for Chemistry Education: To What Extent Can Molecular Visualization and Three-Dimensional Simulations Enhance Meaningful Chemistry Learning? World J. Chem. Educ. 2017, 5, 136–141. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrödinger. Schrödinger Release 2020-4; Maestro, Schrödinger, LLC: New York, NY, USA, 2020. [Google Scholar]
- Pinzi, L.; Rastelli, G. Molecular Docking: Shifting Paradigms in Drug Discovery. Int. J. Mol. Sci. 2019, 20, 4331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Muzio, E.; Toti, D.; Polticelli, F. DockingApp: A User Friendly Interface for Facilitated Docking Simulations with AutoDock Vina. J. Comput. Aided. Mol. Des. 2017, 31, 213–218. [Google Scholar] [CrossRef]
- Cosconati, S.; Forli, S.; Perryman, A.L.; Harris, R.; Goodsell, D.S.; Olson, A.J. Virtual Screening with AutoDock: Theory and Practice. Expert Opin. Drug Discov. 2010, 5, 597–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Case, D.; Ben-Shalom, I.; Brozell, S.; Cerutti, D.; Cheatham, T., III; Cruzeiro, V.; Darden, T.; Duke, R.; Ghoreishi, D.; Gilson, M.; et al. AMBER 18; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Roe, D.R.; Cheatham, T.E. Parallelization of CPPTRAJ Enables Large Scale Analysis of Molecular Dynamics Trajectory Data. J. Comput. Chem. 2018, 39, 2110–2117. [Google Scholar] [CrossRef]
- Wang, E.; Sun, H.; Wang, J.; Wang, Z.; Liu, H.; Zhang, J.Z.H.; Hou, T. End-Point Binding Free Energy Calculation with MM/PBSA and MM/GBSA: Strategies and Applications in Drug Design. Chem. Rev. 2019, 119, 9478–9508. [Google Scholar] [CrossRef]
- Sahoo, B.R.; Basu, M.; Swain, B.; Dikhit, M.R.; Jayasankar, P.; Samanta, M. Elucidation of Novel Structural Scaffold in Rohu TLR2 and Its Binding Site Analysis with Peptidoglycan, Lipoteichoic Acid and Zymosan Ligands, and Downstream MyD88 Adaptor Protein. BioMed Res. Int. 2013, 2013, 185282. [Google Scholar] [CrossRef]
- Sahoo, B.R. Structure of Fish Toll-like Receptors (TLR) and NOD-like Receptors (NLR). Int. J. Biol. Macromol. 2020, 161, 1602. [Google Scholar] [CrossRef]
- Leo, A.; Hansch, C.; Elkins, D. Partition Coefficients and Their Uses. Chem. Rev. 1971, 71, 525–616. [Google Scholar] [CrossRef]
- Daina, A.; Michielin, O.; Zoete, V. ILOGP: A Simple, Robust, and Efficient Description of n-Octanol/Water Partition Coefficient for Drug Design Using the GB/SA Approach. J. Chem. Inf. Model. 2014, 54, 3284–3301. [Google Scholar] [CrossRef] [PubMed]
- Daina, A.; Michielin, O.; Zoete, V. SwissADME: A Free Web Tool to Evaluate Pharmacokinetics, Drug-Likeness and Medicinal Chemistry Friendliness of Small Molecules. Sci. Rep. 2017, 7, 42717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waddad, A.Y.; Ramharack, P.; Soliman, M.E.S.; Govender, T. Grafted hyaluronic acid N-acetyl-l-methionine for targeting of LAT1 receptor: In-silico, synthesis and microscale thermophoresis studies. Int. J. Biol. Macromol. 2019, 125, 767–777. [Google Scholar] [CrossRef]
- Kimuda, M.P.; Laming, D.; Hoppe, H.C.; Tastan Bishop, Ö. Identification of Novel Potential Inhibitors of Pteridine Reductase 1 in Trypanosoma brucei via Computational Structure-Based Approaches and in Vitro Inhibition Assays. Molecules 2019, 24, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavasotto, C.N.; Ortiz, M.A.; Abagyan, R.A.; Piedrafita, F.J. In silico identification of novel EGFR inhibitors with antiproliferative activity against cancer cells. Bioorg. Med. Chem. Lett. 2006, 16, 1969–1974. [Google Scholar] [CrossRef] [PubMed]
- Bollini, M.; Leal, E.S.; Adler, N.S.; Aucar, M.G.; Fernández, G.A.; Pascual, M.J.; Merwaiss, F.; Alvarez, D.E.; Cavasotto, C.N. Discovery of Novel Bovine Viral Diarrhea Inhibitors Using Structure-Based Virtual Screening on the Envelope Protein E2. Front. Chem. 2018, 6, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]







| Peptide Bound to Lipoteichoic Acid | Binding Affinity | |||
|---|---|---|---|---|
BTp1: CTLNGV | Mode | Affinity (kcal/mol) | Dist from rmsd l.b. | Best mode rmsd u.b. |
| 1 | −2.5 | 0 | 0 | |
| 2 | −2.4 | 9.222 | 12.669 | |
| 3 | −2.3 | 5.016 | 7.706 | |
| 4 | −2.2 | 8.403 | 13.391 | |
| 5 | −2.2 | 8.094 | 12.23 | |
| 6 | −2.2 | 9.072 | 14.517 | |
| 7 | −2.2 | 9.339 | 13.51 | |
| 8 | −2.2 | 8.196 | 11.353 | |
| 9 | −2.1 | 2.631 | 8.354 | |
BTp2: RRLHIPRF | Mode | Affinity (Kcal/mol) | Dist from rmsd l.b. | Best mode rmsd u.b. |
| 1 | −3.4 | 0 | 0 | |
| 2 | −3.4 | 3.659 | 10.855 | |
| 3 | −3.4 | 2.62 | 5.43 | |
| 4 | −3.3 | 1.369 | 2.413 | |
| 5 | −3.3 | 5.024 | 9.196 | |
| 6 | −3.2 | 3.682 | 11.033 | |
| 7 | −3.2 | 4.461 | 10.666 | |
| 8 | −3.2 | 1.816 | 3.079 | |
| 9 | −3.2 | 5.128 | 8.731 | |
BTp3: YDLLYSLT | Mode | Affinity (Kcal/mol) | Dist from rmsd l.b. | Best mode rmsd u.b. |
| 1 | −2.9 | 0 | 0 | |
| 2 | −2.8 | 2.658 | 5.748 | |
| 3 | −2.8 | 1.639 | 4.576 | |
| 4 | −2.7 | 4.626 | 9.394 | |
| 5 | −2.7 | 8.39 | 12.838 | |
| 6 | −2.7 | 5.11 | 9.703 | |
| 7 | −2.7 | 8.117 | 12.937 | |
| 8 | −2.6 | 3.682 | 8.654 | |
| 9 | −2.6 | 4.175 | 8.33 | |
BTp4: SKVFLVP | Mode | Affinity (Kcal/mol) | Dist from rmsd l.b. | Best mode rmsd u.b. |
| 1 | −3.8 | 0 | 0 | |
| 2 | −3.7 | 3.574 | 6.868 | |
| 3 | −3.7 | 1.862 | 2.555 | |
| 4 | −3.6 | 2.005 | 3.056 | |
| 5 | −3.6 | 2.577 | 3.93 | |
| 6 | −3.6 | 3.501 | 7.01 | |
| 7 | −3.5 | 2.83 | 6.586 | |
| 8 | −3.5 | 7.209 | 12.029 | |
| 9 | −3.4 | 1.885 | 3.139 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Devnarain, N.; Waddad, A.Y.; de la Torre, B.G.; Albericio, F.; Govender, T. Novel Biomimetic Human TLR2-Derived Peptides for Potential Targeting of Lipoteichoic Acid: An In Silico Assessment. Biomedicines 2021, 9, 1063. https://doi.org/10.3390/biomedicines9081063
Devnarain N, Waddad AY, de la Torre BG, Albericio F, Govender T. Novel Biomimetic Human TLR2-Derived Peptides for Potential Targeting of Lipoteichoic Acid: An In Silico Assessment. Biomedicines. 2021; 9(8):1063. https://doi.org/10.3390/biomedicines9081063
Chicago/Turabian StyleDevnarain, Nikita, Ayman Y. Waddad, Beatriz G. de la Torre, Fernando Albericio, and Thirumala Govender. 2021. "Novel Biomimetic Human TLR2-Derived Peptides for Potential Targeting of Lipoteichoic Acid: An In Silico Assessment" Biomedicines 9, no. 8: 1063. https://doi.org/10.3390/biomedicines9081063
APA StyleDevnarain, N., Waddad, A. Y., de la Torre, B. G., Albericio, F., & Govender, T. (2021). Novel Biomimetic Human TLR2-Derived Peptides for Potential Targeting of Lipoteichoic Acid: An In Silico Assessment. Biomedicines, 9(8), 1063. https://doi.org/10.3390/biomedicines9081063