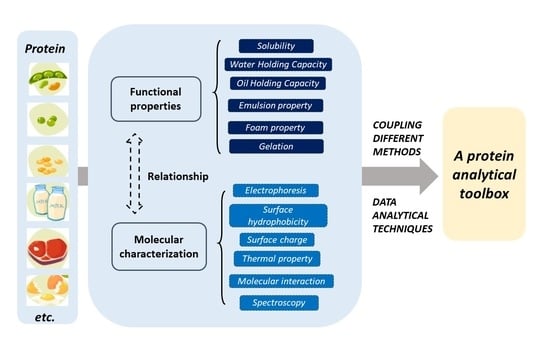
Survey on Methods for Investigating Protein Functionality and Related Molecular Characteristics
Abstract
:1. Introduction
2. Protein Functionality Methods
2.1. Solubility
2.2. Water Holding Capacity (WHC)
2.3. Oil Holding Capacity (OHC)
2.4. Foam Properties
2.5. Emulsion Properties
2.6. Gelation
3. Molecular Characterization Methods
3.1. Electrophoresis
3.2. Surface Hydrophobicity (So) and Surface Charge
3.3. Thermal Property
3.4. Molecular Interactions
3.5. Spectroscopy
3.5.1. Ultraviolet-Visible (UV-Vis) Spectroscopy
3.5.2. Fourier Transform Infrared (FTIR) Spectroscopy
3.5.3. Raman Spectroscopy
3.5.4. Circular Dichroism (CD) Spectroscopy
3.5.5. Fluorescence Spectroscopy
3.5.6. Nuclear Magnetic Resonance (NMR) Spectroscopy
4. Relationship between Structural and Functionality Features
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Bessada, S.M.F.; Barreira, J.C.M.; Oliveira, M.B.P.P. Pulses and food security: Dietary protein, digestibility, bioactive and functional properties. Trends Food Sci. Technol. 2019, 93, 53–68. [Google Scholar] [CrossRef]
- Boye, J.; Zare, F.; Pletch, A. Pulse proteins: Processing, characterization, functional properties and applications in food and feed. Food Res. Int. 2010, 43, 414–431. [Google Scholar] [CrossRef]
- Day, L. Proteins from land plants—Potential resources for human nutrition and food security. Trends Food Sci. Technol. 2013, 32, 25–42. [Google Scholar] [CrossRef]
- Sim, S.Y.J.; Srv, A.; Chiang, J.H.; Henry, C.J. Plant Proteins for Future Foods: A Roadmap. Foods 2021, 10, 1967. [Google Scholar] [CrossRef]
- Nakai, S. Structure-function relationships of food proteins: With an emphasis on the importance of protein hydrophobicity. J. Agric. Food Chem. 1983, 31, 676–683. [Google Scholar] [CrossRef]
- Mune Mune, M.A.; Sogi, D.S.; Minka, S.R. Response surface methodology for investigating structure–function relationship of grain legume proteins. J. Food Process. Preserv. 2018, 42, e13524. [Google Scholar] [CrossRef]
- Shevkani, K.; Singh, N.; Kaur, A.; Rana, J.C. Structural and functional characterization of kidney bean and field pea protein isolates: A comparative study. Food Hydrocoll. 2015, 43, 679–689. [Google Scholar] [CrossRef]
- Wang, K.; Sun, D.-W.; Pu, H.; Wei, Q. Principles and applications of spectroscopic techniques for evaluating food protein conformational changes: A review. Trends Food Sci. Technol. 2017, 67, 207–219. [Google Scholar] [CrossRef]
- Zayas, J.F. Functionality of Proteins in Food, 1st ed.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 1–4. [Google Scholar]
- Li-Chan, E.C.Y.; Lacroix, I.M.E. 1—Properties of proteins in food systems: An introduction. In Proteins in Food Processing, 2nd ed.; Yada, R.Y., Ed.; Woodhead Publishing: Cambridge, UK, 2018; pp. 1–25. [Google Scholar]
- Culbertson, J. Food Protein Functionality. In Handbook of Food Science, Technology, and Engineering, 1st ed.; Hui, Y., Sherkat, F., Eds.; CRC Press: Boca Raton, FL, USA, 2005; Volume 1, pp. 1–12. [Google Scholar]
- Sathe, S.K.; Zaffran, V.D.; Gupta, S.; Li, T. Protein Solubilization. J. Amer. Oil Chem. Soc. 2018, 95, 883–901. [Google Scholar] [CrossRef]
- Morr, C.V.; German, B.; Kinsella, J.E.; Regenstein, J.M.; Buren, J.P.V.; Kilara, A.; Lewis, B.A.; Mangino, M.E. A collaborative study to develop a standardized food protein solubility procedure. J. Food Sci. 1985, 50, 1715–1718. [Google Scholar] [CrossRef]
- Mæhre, H.K.; Dalheim, L.; Edvinsen, G.K.; Elvevoll, E.O.; Jensen, I.-J. Protein determination—method matters. Foods 2018, 7, 5. [Google Scholar] [CrossRef] [Green Version]
- Kramer, R.M.; Shende, V.R.; Motl, N.; Pace, C.N.; Scholtz, J.M. Toward a molecular understanding of protein solubility: Increased negative surface charge correlates with increased solubility. Biophys. J. 2012, 102, 1907–1915. [Google Scholar] [CrossRef] [Green Version]
- Adebiyi, A.P.; Aluko, R.E. Functional properties of protein fractions obtained from commercial yellow field pea (Pisum sativum L.) seed protein isolate. Food Chem. 2011, 128, 902–908. [Google Scholar] [CrossRef]
- Elsohaimy, S.A.; Refaay, T.M.; Zaytoun, M.A.M. Physicochemical and functional properties of quinoa protein isolate. Ann. Agric. Sci. 2015, 60, 297–305. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Shen, C.; Wu, Z.; Zhang, Z.; Xu, C. Comparison of wheat, soybean, rice, and pea protein properties for effective applications in food products. J. Food Biochem. 2020, 44, e13157. [Google Scholar] [CrossRef]
- Hermansson, A. Water and fat holding. In Functional Properties of Food Macromolecules, 1st ed.; Mitchell, J.R., Ledward, D.A., Eds.; Elsevier Applied Science: London, UK, 1986; pp. 273–314. [Google Scholar]
- Zayas, J.F. Water Holding Capacity of Proteins. In Functionality of Proteins in Food, 1st ed.; Zayas, J.F., Ed.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 76–133. [Google Scholar]
- Quinn, J.R.; Paton, D. A practical measurement of water hydration capacity of protein materials. Cereal Chem. 1979, 56, 38–40. [Google Scholar]
- Liu, Y.; Wang, D.; Wang, J.; Yang, Y.; Zhang, L.; Li, J.; Wang, S. Functional properties and structural characteristics of phosphorylated pea protein isolate. Int. J. Food Sci. Technol. 2020, 55, 2002–2010. [Google Scholar] [CrossRef]
- Alu’Datt, M.H.; Rababah, T.; Alhamad, M.N.; Ereifej, K.; Gammoh, S.; Kubow, S.; Tawalbeh, D. Preparation of mayonnaise from extracted plant protein isolates of chickpea, broad bean and lupin flour: Chemical, physiochemical, nutritional and therapeutic properties. J. Food Sci. Technol. 2017, 54, 1395–1405. [Google Scholar] [CrossRef] [Green Version]
- Bühler, J.M.; Dekkers, B.L.; Bruins, M.E.; Van Der Goot, A.J. Modifying faba bean protein concentrate using dry heat to increase water holding capacity. Foods 2020, 9, 1077. [Google Scholar] [CrossRef]
- Xu, Y.; Obielodan, M.; Sismour, E.; Arnett, A.; Alzahrani, S.; Zhang, B. Physicochemical, functional, thermal and structural properties of isolated Kabuli chickpea proteins as affected by processing approaches. Int. J. Food Sci. 2017, 52, 1147–1154. [Google Scholar] [CrossRef]
- Zayas, J.F. Oil and fat binding properties of proteins. In Functionality of Proteins in Food, 1st ed.; Zayas, J.F., Ed.; Springer: Berlin/Heidelberg, Germany, 1997; pp. 228–259. [Google Scholar]
- Kinsella, J.E.; Melachouris, N. Functional properties of proteins in foods: A survey. Crit. Rev. Food Sci. Nutr. 1976, 7, 219–280. [Google Scholar] [CrossRef]
- Kinsella, J.E. Functional properties of soy proteins. J. Am. Oil Chem. Soc. 1979, 56, 242–258. [Google Scholar] [CrossRef]
- Sathe, S.K.; Deshpande, S.S.; Salunkhe, D.K. Functional properties of winged bean [Psophocarpus tetragonolobus (L.) DC] proteins. J. Food Sci. 1982, 47, 503–509. [Google Scholar] [CrossRef]
- Lam, A.C.Y.; Warkentin, T.D.; Tyler, R.T.; Nickerson, M.T. Physicochemical and functional properties of protein isolates obtained from several pea cultivars. Cereal Chem. 2017, 94, 89–97. [Google Scholar] [CrossRef]
- Sharan, S.; Zanghelini, G.; Zotzel, J.; Bonerz, D.; Aschoff, J.; Saint-Eve, A.; Maillard, M.N. Fava bean (Vicia faba L.) for food applications: From seed to ingredient processing and its effect on functional properties, antinutritional factors, flavor, and color. Compr. Rev. Food Sci. Food Saf. 2021, 20, 401–428. [Google Scholar] [CrossRef]
- Multari, S.; Stewart, D.; Russell, W. Potential of fava bean as future protein supply to partially replace meat intake in the human diet. Compr. Rev. Food Sci. Food Saf. 2015, 14, 511–522. [Google Scholar] [CrossRef]
- Arogundade, L.A.; Tshay, M.; Shumey, D.; Manazie, S. Effect of ionic strength and/or pH on extractability and physico-functional characterization of broad bean (Vicia faba L.) protein concentrate. Food Hydrocoll. 2006, 20, 1124–1134. [Google Scholar] [CrossRef]
- Makri, E.; Papalamprou, E.; Doxastakis, G. Study of functional properties of seed storage proteins from indigenous European legume crops (lupin, pea, broad bean) in admixture with polysaccharides. Food Hydrocoll. 2005, 19, 583–594. [Google Scholar] [CrossRef]
- Sosulski, F.W.; McCurdy, A. Functionality of flours, protein factions and isolates from field peas and faba bean. J. Food Sci. 1987, 52, 1010–1014. [Google Scholar] [CrossRef]
- Paredes-López, O.; Ordorica-Falomir, C.; Olivares-Vázquez, M.R. Chickpea protein isolates: Physicochemical, functional and nutritional characterization. J. Food Sci. 1991, 56, 726–729. [Google Scholar] [CrossRef]
- Gao, W.-R.; Wang, X.-S.; Li, J.-G.; Zhang, J.-S.; Ma, H. Physicochemical and processing functional properties of proteins from two chinese chickpea (Cicer arietinum L.) Cultivars. J. Food Process. Preserv. 2009, 34, 575–594. [Google Scholar]
- Husband, F.A.; Wilde, P.J.; Clark, D.C.; Rawel, H.M.; Muschiolik, G. Foaming properties of modified faba bean protein isolates. Food Hydrocoll. 1994, 8, 455–468. [Google Scholar] [CrossRef]
- Chao, D.; Aluko, R.E. Modification of the structural, emulsifying, and foaming properties of an isolated pea protein by thermal pretreatment. CYTA J. Food 2018, 16, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Damodaran, S. Protein Stabilization of Emulsions and Foams. J. Food Sci. 2005, 70, 54–66. [Google Scholar] [CrossRef]
- Felix, M.; Lopez-Osorio, A.; Romero, A.; Guerrero, A. Faba bean protein flour obtained by densification: A sustainable method to develop protein concentrates with food applications. LWT Food Sci. Technol. 2018, 93, 563–569. [Google Scholar] [CrossRef]
- Burger, T.G.; Zhang, Y. Recent progress in the utilization of pea protein as an emulsifier for food applications. Trends Food Sci. Technol. 2019, 86, 25–33. [Google Scholar] [CrossRef]
- Stone, A.K.; Karalash, A.; Tyler, R.T.; Warkentin, T.D.; Nickerson, M.T. Functional attributes of pea protein isolates prepared using different extraction methods and cultivars. Food Res. Int. 2015, 76, 31–38. [Google Scholar] [CrossRef]
- Yasumatsu, K.; Sawada, K.; Moritaka, S.; Misaki, M.; Toda, J.; Wada, T.; Ishii, K. Whipping and emulsifying properties of soybean products. Agric. Biol. Chem. 1972, 36, 719–727. [Google Scholar] [CrossRef]
- Chao, D.; Jung, S.; Aluko, R.E. Physicochemical and functional properties of high pressure-treated isolated pea protein. Innov. Food Sci. Emerg. Technol. 2018, 45, 179–185. [Google Scholar] [CrossRef]
- Cui, L.; Bandillo, N.; Wang, Y.; Ohm, J.-B.; Chen, B.; Rao, J. Functionality and structure of yellow pea protein isolate as affected by cultivars and extraction pH. Food Hydrocoll. 2020, 108, 106008. [Google Scholar] [CrossRef]
- Ge, J.; Sun, C.X.; Corke, H.; Gul, K.; Gan, R.Y.; Fang, Y. The health benefits, functional properties, modifications, and applications of pea (Pisum sativum L.) protein: Current status, challenges, and perspectives. Compr. Rev. Food Sci. Food Saf. 2020, 19, 1835–1876. [Google Scholar] [CrossRef]
- Dakhili, S.; Abdolalizadeh, L.; Hosseini, S.M.; Shojaee-Aliabadi, S.; Mirmoghtadaie, L. Quinoa protein: Composition, structure and functional properties. Food Chem. 2019, 299, 125161. [Google Scholar] [CrossRef]
- Chmielewska, A.; Kozłowska, M.; Rachwał, D.; Wnukowski, P.; Amarowicz, R.; Nebesny, E.; Rosicka-Kaczmarek, J. Canola/rapeseed protein—nutritional value, functionality and food application: A review. Crit. Rev. Food Sci. Nutr. 2020. [Google Scholar] [CrossRef]
- Wang, X.; Gao, W.; Zhang, J.; Zhang, H.; Li, J.; He, X.; Ma, H. Subunit, amino acid composition and in vitro digestibility of protein isolates from Chinese kabuli and desi chickpea (Cicer arietinum L.) cultivars. Food Res. Int. 2010, 43, 567–572. [Google Scholar] [CrossRef]
- Sun, X.D.; Arntfield, S.D. Gelation properties of salt-extracted pea protein isolate induced by heat treatment: Effect of heating and cooling rate. Food Chem. 2011, 124, 1011–1016. [Google Scholar] [CrossRef]
- Manns, J.M. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) of proteins. Curr. Protoc. Microbiol. 2011, 22, A.3M.1–A.3M.13. [Google Scholar] [CrossRef]
- Murphy, K.P.; Privalov, P.L.; Gill, S.J. Common features of protein unfolding and dissolution of hydrophobic compounds. Science 1990, 247, 559–561. [Google Scholar] [CrossRef]
- Cardamone, M.; Puri, N.K. Spectrofluorimetric assessment of the surface hydrophobicity of proteins. Biochem. J. 1992, 282, 589–593. [Google Scholar] [CrossRef]
- Nakai, S.; Li-Chan, E.; Arteaga, G. Measurement of surface hydrophobicity. In Methods of Testing Protein Functionality, 1st ed.; Hall, G.M., Ed.; Chapman & Hall: London, UK, 1996; Volume 17, pp. 226–259. [Google Scholar]
- He, X.; Chen, J.; He, X.; Feng, Z.; Li, C.; Liu, W.; Dai, T.; Liu, C. Industry-scale microfluidization as a potential technique to improve solubility and modify structure of pea protein. Innov. Food Sci. Emerg. Technol. 2021, 67, 102582. [Google Scholar] [CrossRef]
- Xiong, T.; Xiong, W.; Ge, M.; Xia, J.; Li, B.; Chen, Y. Effect of high intensity ultrasound on structure and foaming properties of pea protein isolate. Food Res. Int. 2018, 109, 260–267. [Google Scholar] [CrossRef]
- Fang, L.; Xiang, H.; Sun-Waterhouse, D.; Cui, C.; Lin, J. Enhancing the Usability of Pea Protein Isolate in Food Applications through Modifying Its Structural and Sensory Properties via Deamidation by Glutaminase. J. Agric. Food Chem. 2020, 68, 1691–1697. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Shen, Y.; Zhang, Y.; Schilling, M.W.; Li, Y. Parallel comparison of functional and physicochemical properties of common pulse proteins. LWT Food Sci. Technol. 2021, 146, 111594. [Google Scholar] [CrossRef]
- Grossmann, L.; Kinchla, A.J.; Nolden, A.; McClements, D.J. Standardized methods for testing the quality attributes of plant-based foods: Milk and cream alternatives. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2206–2233. [Google Scholar] [CrossRef] [PubMed]
- Gill, P.; Moghadam, T.T.; Ranjbar, B. Differential scanning calorimetry techniques: Applications in biology and nanoscience. J. Biomol. Tech. 2010, 21, 167–193. [Google Scholar]
- Seenivasan, A.; Panda, T. Protein characterization by thermal property measurement. In Glass Transition and Phase Transitions in Food and Biological Materials, 1st ed.; Ahmed, J., Rahman, M.S., Roosiley, Y.H., Eds.; John Wiley & Sons Ltd.: Chichester, UK, 2017; pp. 305–391. [Google Scholar]
- Puppo, C.; Chapleau, N.; Speroni, F.; De Lamballerie-Anton, M.; Michel, F.; Añón, C.; Anton, M. Physicochemical modifications of high-pressure-treated soybean protein isolates. J. Agric. Food Chem. 2004, 52, 1564–1571. [Google Scholar] [CrossRef]
- Arntfield, S.D.; Murray, E.D. The influence of processing parameters on food protein functionality I. differential scanning calorimetry as an indicator of protein denaturation. Can. Inst. Food Technol. J. 1981, 14, 289–294. [Google Scholar] [CrossRef]
- Liu, K.; Hsieh, F.-H. Protein–protein interactions during high-moisture extrusion for fibrous meat analogues and comparison of protein solubility methods using different solvent systems. J. Agric. Food Chem. 2008, 56, 2681–2687. [Google Scholar] [CrossRef]
- Chen, F.L.; Wei, Y.M.; Zhang, B. Chemical cross-linking and molecular aggregation of soybean protein during extrusion cooking at low and high moisture content. LWT Food Sci. Technol. 2011, 44, 957–962. [Google Scholar] [CrossRef]
- Malik, A.K.; Kumar, R.; Heena. Spectroscopy: Types. In Encyclopedia of Food and Health, 1st ed.; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; pp. 64–72. [Google Scholar]
- Antosiewicz, J.M.; Shugar, D. UV–Vis spectroscopy of tyrosine side-groups in studies of protein structure. Part 2: Selected applications. Biophys. Rev. 2016, 8, 163–177. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, T.; He, F.; Chen, G. Fabrication of pea protein-curcumin nanocomplexes via microfluidization for improved solubility, nano-dispersibility and heat stability of curcumin: Insight on interaction mechanisms. Int. J. Biol. Macromol. 2021, 168, 686–694. [Google Scholar] [CrossRef]
- Sha, L.; Koosis, A.O.; Wang, Q.; True, A.D.; Xiong, Y.L. Interfacial dilatational and emulsifying properties of ultrasound-treated pea protein. Food Chem. 2021, 350, 129271. [Google Scholar] [CrossRef]
- Hansen, S.K.; Jamali, B.; Hubbuch, J. Selective high throughput protein quantification based on UV absorption spectra. Biotechnol. Bioeng. 2013, 110, 448–460. [Google Scholar] [CrossRef]
- Lin, M.; Rasco, B.A.; Cavinato, A.G.; Al-Holy, M. Chapter 6—Infrared (IR) Spectroscopy—Near-Infrared Spectroscopy and Mid-Infrared Spectroscopy. In Infrared Spectroscopy for Food Quality Analysis and Control, 1st ed.; Sun, D.-W., Ed.; Academic Press: Amsterdam, The Netherlands, 2009; pp. 119–143. [Google Scholar]
- Kong, J.; Yu, S. Fourier transform infrared spectroscopic analysis of protein secondary structures. Acta Biochim. Biophys. Sin. 2007, 39, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Carbonaro, M.; Nucara, A. Secondary structure of food proteins by Fourier transform spectroscopy in the mid-infrared region. Amino Acids 2010, 38, 679–690. [Google Scholar] [CrossRef]
- Beck, S.M.; Knoerzer, K.; Arcot, J. Effect of low moisture extrusion on a pea protein isolate’s expansion, solubility, molecular weight distribution and secondary structure as determined by Fourier Transform Infrared Spectroscopy (FTIR). J. Food Eng. 2017, 214, 166–174. [Google Scholar] [CrossRef]
- Jin, H.; Lu, Q.; Chen, X.; Ding, H.; Gao, H.; Jin, S. The use of Raman spectroscopy in food processes: A review. Appl. Spectrosc. Rev. 2016, 51, 12–22. [Google Scholar] [CrossRef]
- Yang, D.; Ying, Y. Applications of Raman Spectroscopy in Agricultural Products and Food Analysis: A Review. Appl. Spectrosc. Rev. 2011, 46, 539–560. [Google Scholar] [CrossRef]
- Rygula, A.; Majzner, K.; Marzec, K.M.; Kaczor, A.; Pilarczyk, M.; Baranska, M. Raman spectroscopy of proteins: A review. J. Raman Spectrosc. 2013, 44, 1061–1076. [Google Scholar] [CrossRef]
- Wei, D.; Chen, S.; Liu, Q. Review of Fluorescence Suppression Techniques in Raman Spectroscopy. Appl. Spectrosc. Rev. 2015, 50, 387–406. [Google Scholar] [CrossRef]
- Weng, S.; Zhu, W.; Zhang, X.; Yuan, H.; Zheng, L.; Zhao, J.; Huang, L.; Han, P. Recent advances in Raman technology with applications in agriculture, food and biosystems: A review. Artif. Intell. Agric. 2019, 3, 1–10. [Google Scholar] [CrossRef]
- Kelly, S.M.; Jess, T.J.; Price, N.C. How to study proteins by circular dichroism. BBA Proteins Proteom. 2005, 1751, 119–139. [Google Scholar] [CrossRef]
- Kelly, S.; Price, N. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 2000, 1, 349–384. [Google Scholar] [CrossRef] [Green Version]
- Whitmore, L.; Wallace, B.A. Protein secondary structure analyses from circular dichroism spectroscopy: Methods and reference databases. Biopolymers 2008, 89, 392–400. [Google Scholar] [CrossRef]
- Martin, S.R.; Schilstra, M.J. Circular dichroism and its application to the study of biomolecules. Methods Cell Biol. 2008, 84, 263–293. [Google Scholar]
- Miles, A.J.; Janes, R.W.; Wallace, B.A. Tools and methods for circular dichroism spectroscopy of proteins: A tutorial review. Chem. Soc. Rev. 2021, 50, 8400–8413. [Google Scholar] [CrossRef]
- Faassen, S.; Hitzmann, B. Fluorescence Spectroscopy and Chemometric Modeling for Bioprocess Monitoring. Sensors 2015, 15, 10271–10291. [Google Scholar] [CrossRef] [Green Version]
- Sikorska, E.; Khmelinskii, I.; Sikorski, M. Fluorescence spectroscopy and imaging instruments for food quality evaluation. In Evaluation Technologies for Food Quality, 1st ed.; Zhong, J., Wang, X., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 491–533. [Google Scholar]
- Lakowicz, J.R. Principles of Fluorescence Spectroscopy, 3rd ed.; Springer: Boston, MA, USA, 2006; pp. 63–66. [Google Scholar]
- Royer, C.A. Probing Protein Folding and Conformational Transitions with Fluorescence. Chem. Rev. 2006, 106, 1769–1784. [Google Scholar] [CrossRef]
- SádeCká, J.; TóThoVá, J. Fluorescence spectroscopy and chemometrics in the food classification—A review. Czech J. Food Sci. 2007, 25, 159–173. [Google Scholar] [CrossRef] [Green Version]
- Dankowska, A. Advances in Fluorescence Emission Spectroscopy for Food Authenticity Testing. In Advances in Food Authenticity Testing, 1st ed.; Downey, G., Ed.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 117–145. [Google Scholar]
- Tian, Y.; He, Q.; Chen, X.; Wang, S. Nuclear magnetic resonance spectroscopy for food quality evaluation. In Evaluation Technologies for Food Quality, 1st ed.; Zhong, J., Wang, X., Eds.; Elsevier: Amsterdam, The Netherlands, 2019; pp. 193–217. [Google Scholar]
- Hatzakis, E. Nuclear Magnetic Resonance (NMR) Spectroscopy in Food Science: A Comprehensive Review. Compr. Rev. Food Sci. Food Saf. 2019, 18, 189–220. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, A.; Mitsuiki, M.; Motoki, M.; Ebisawa, K.; Suzuki, E.-I. Relationship between the Glass Transition of Soy Protein and Molecular Structure. J. Agric. Food Chem. 2000, 48, 3292–3297. [Google Scholar] [CrossRef]
- Kaas, Q.; Craik, D.J. NMR of plant proteins. Prog. Nucl. Magn. Reson. Spectrosc. 2013, 71, 1–34. [Google Scholar] [CrossRef] [PubMed]
- Avramenko, N.A.; Low, N.H.; Nickerson, M.T. The effects of limited enzymatic hydrolysis on the physicochemical and emulsifying properties of a lentil protein isolate. Food Res. Int. 2013, 51, 162–169. [Google Scholar] [CrossRef]
- Sharan, S.; Zotzel, J.; Stadtmüller, J.; Bonerz, D.; Aschoff, J.; Saint-Eve, A.; Maillard, M.-N.; Olsen, K.; Rinnan, Å.; Orlien, V. Two Statistical Tools for Assessing Functionality and Protein Characteristics of Different Fava Bean (Vicia faba L.) Ingredients. Foods 2021, 10, 2489. [Google Scholar] [CrossRef] [PubMed]
- Kadiroğlu, P.; Aydemir, L.Y.; Akcakaya, F.G. Prediction of functional properties of registered chickpea samples using FT-IR spectroscopy and chemometrics. LWT Food Sci. Technol. 2018, 93, 463–469. [Google Scholar] [CrossRef]
- Keivaninahr, F.; Gadkari, P.; Zoroufchi Benis, K.; Tulbek, M.; Ghosh, S. Prediction of emulsification behaviour of pea and faba bean protein concentrates and isolates from structure–functionality analysis. RSC Adv. 2021, 11, 12117–12135. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Sharan, S.; Rinnan, Å.; Orlien, V. Survey on Methods for Investigating Protein Functionality and Related Molecular Characteristics. Foods 2021, 10, 2848. https://doi.org/10.3390/foods10112848
Zhang Y, Sharan S, Rinnan Å, Orlien V. Survey on Methods for Investigating Protein Functionality and Related Molecular Characteristics. Foods. 2021; 10(11):2848. https://doi.org/10.3390/foods10112848
Chicago/Turabian StyleZhang, Yuqi, Siddharth Sharan, Åsmund Rinnan, and Vibeke Orlien. 2021. "Survey on Methods for Investigating Protein Functionality and Related Molecular Characteristics" Foods 10, no. 11: 2848. https://doi.org/10.3390/foods10112848
APA StyleZhang, Y., Sharan, S., Rinnan, Å., & Orlien, V. (2021). Survey on Methods for Investigating Protein Functionality and Related Molecular Characteristics. Foods, 10(11), 2848. https://doi.org/10.3390/foods10112848