Impact of Gastrointestinal Digestion Simulation on the Formation of Angiotensin-I-Converting Enzyme Inhibitory (ACE-I) Peptides from Germinated Lamtoro Gung Flour
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
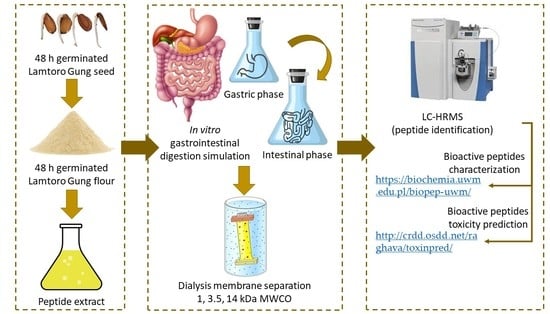
2.2. Peptide Extraction
2.3. GID Simulation
2.4. Degree of Hydrolysis (DH), Peptide Concentration Assay, and the MW Distribution Calculation
2.5. ACE-I Activity Assay
2.6. Peptide Fractionation Using the MW Cut Off (MWCO) Filtration
2.7. Amino Acid Composition
2.8. Characterisation of the Peptides and the Sequence Identification
2.9. Biological Potential of the Peptides
2.10. Statistical Analysis
3. Results
3.1. %DH and the Peptide Concentration of the Germinated Lamtoro Gung Flour during the GID Simulation
3.2. ACE-I Activity of the Germinated Lamtoro Gung Flour during the GID Simulation
3.3. Peptide Fractionation
3.3.1. Peptide Concentration of Each Fraction
3.3.2. MW Distribution of Each Fraction
3.3.3. ACE-I Activity of Each Peptide Fraction
3.3.4. Amino Acid Composition
3.3.5. Identification of the Lamtoro Gung Flour Peptide and its Inhibitory Effect
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verma, S. A Review Study on Leucaena leucocephala: A Multipurpose Tree. Int. J. Sci. Res. Sci. Eng. Technol. 2016, 2, 103–105. [Google Scholar]
- Harifah, C.S. Perubahan Zat Gizi, Senyawa Antigizi, serta nilai Cerna Protein secara In Vitro serta Profil Asam Amino Biji Lamtoro Gung (Leucaena leucocephala) Kukus dan Rebus. Master’s Thesis, Universitas Gadjah Mada, Sleman, Special District of Yogyakarta, Indonesia, 2017. [Google Scholar]
- Supriyadi, S.; Indrati, R.; Santoso, U. Peptida Bioaktif dari Indigenous Indonesian Stinky Bean sebagai Sumber ACE-Inhibitor untuk Menekan Penyakit Hipertensi. Lap. PTUPT 2021 2021, 2, 1–3. [Google Scholar]
- Thewissen, B.G.; Pauly, A.; Celus, I.; Brijs, K.; Delcour, J.A. Inhibition of angiotensin I-converting enzyme by wheat gliadin hydrolysates. Food Chem. 2011, 127, 1653–1658. [Google Scholar] [CrossRef]
- Rui, X.; Boye, J.I.; Simpson, B.K.; Prasher, S.O. Purification and Characterization of Cngiotensin I-converting Enzyme Inhibitory Peptides of Small Red Bean (Phaseolus vulgaris) Hydrolysates. J. Funct. Foods 2013, 5, 1116–1124. [Google Scholar] [CrossRef]
- Aluko, R.E. Structure and function of plant protein-derived antihypertensive peptides. Curr. Opin. Food Sci. 2015, 4, 44–50. [Google Scholar] [CrossRef]
- Fan, H.; Liao, W.; Wu, J. Molecular Interactions, Bioavailability, and Cellular Mechanisms of Angiotensin-converting Enzyme Inhibitory Peptides. J. Food Biochem. 2018, 43, e12572. [Google Scholar] [CrossRef] [Green Version]
- Natesh, R.; Schwager SL, U.; Sturrock, E.D.; Acharya, K.R. Crystal Structure of The Human Angiotensin-converting Enzyme–Lisinopril Complex. Nat. Publ. Gr. 2003, 421, 551–554. [Google Scholar] [CrossRef] [Green Version]
- Masuyer, G.; Douglas, R.G.; Sturrock, E.D.; Acharya, K.R. Structural basis of Ac-SDKP hydrolysis by Angiotensin-I converting enzyme. Nat. Publ. Gr. 2015, 5, 13742. [Google Scholar] [CrossRef] [Green Version]
- Puspitojati, E.; Cahyanto, M.N.; Marsono, Y.; Indrati, R. Changes in Amino Acid Composition during Fermentation and Its Effects on The Inhibitory Activity of Angiotensin-I-converting Enzyme of Jack Bean Tempe Following In vitro Gastrointestinal Digestion. J. Food Nutr. Res. 2019, 58, 319–327. [Google Scholar]
- Puspitojati, E.; Cahyanto, M.N.; Marsono, Y.; Indrati, R. Production of Angiotensin-I-Converting Enzyme (ACE) Inhibitory Peptides during the Fermentation of Jack Bean (Canavalia ensiformis) Tempe. Pakistan J. Nutr. 2019, 18, 464–470. [Google Scholar] [CrossRef] [Green Version]
- Pedroche, J.; Yust, M.M.; Giron-Calle, J.; Alaiz, M.; Millan, F.; Vioque, J. Utilisation of Chickpea Protein Isolates for Production of Peptides with Angiotensin I-Converting Enzyme (ACE)-Inhibitory Activity. J. Sci. Food Agric. 2002, 965, 960–965. [Google Scholar] [CrossRef]
- Ratnayani, K.; Suter, I.K.; Antara, N.S.; Putra, I.N.K. Angiotensin converting enzyme (ACE) inhibitory activity of peptide fraction of germinated Pigeon Pea (Cajanus cajan (L.) Millsp.). Indones. J. Chem. 2019, 19, 900–906. [Google Scholar] [CrossRef] [Green Version]
- Mamilla, R.K.; Mishra, V.K. Effect of germination on antioxidant and ACE inhibitory activities of legumes. LWT-Food Sci. Technol. 2017, 75, 51–58. [Google Scholar] [CrossRef]
- Bamdad, F.; Dokhani, S.; Keramat, J.; Zareie, R. The impact of germination and in vitro digestion on the formation of Angiotensin converting enzyme (ACE) inhibitory peptides from lentil proteins compared to whey proteins. Int. J. Biol. Life Sci. 2009, 5, 2009. [Google Scholar]
- Ratnayani, K.; Suter, I.K.; Antara, N.S.; Putra, I.N.K. Effect of in vitro Gastrointestinal Digestion on The Angiotensin Converting Enzyme (ACE) Inhibitory Activity of Pigeon Pea Protein Isolate. Int. Food Res. J. 2019, 26, 1397–1404. [Google Scholar]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food-an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Fitriani, A.; Indrati, R.; Marsono, Y.; Supriyadi, S. Angiotensin-I-converting enzyme inhibitory (ACE-I) peptide from germinated Lamtoro Gung (Leucaena laucocephala ssp. Glabrata (Rose) S. Zarate) flour. Sains Malays. 2022, 51, 1–25. [Google Scholar]
- Pertiwi MG, P.; Marsono, Y.; Indrati, R. In Vitro Gastrointestinal Simulation of Tempe Prepared from Koro Kratok (Phaseolus lunatus L.) as An Angiotensin-converting Enzyme Inhibitor. J. Food Sci. Technol. 2019, 57, 1847–1855. [Google Scholar] [CrossRef]
- Charoenphun, N.; Cheirsilp, B.; Sirinupong, N.; Youravong, W. Calcium-binding Peptides Derived from Tilapia (Oreochromis niloticus) Protein Hydrolysate. Eur. Food Res. Technol. 2013, 236, 57–63. [Google Scholar] [CrossRef]
- Minkiewicz, P.; Iwaniak, A.; Darewicz, M. BIOPEP-UWM database of bioactive peptides: Current opportunities. Int. J. Mol. Sci. 2019, 20, 5978. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Kapoor, P.; Chaudhary, K.; Gautam, A.; Kumar, R.; Raghava, G.P.S. In Silico Approach for Predicting Toxicity of Peptides and Proteins. PLoS ONE 2013, 8, e73957. [Google Scholar] [CrossRef] [Green Version]
- Berdutina, A.V.; Neklyudov, A.D.; Ivankin, A.I.; Karpo, B.S.; Mitaleva, S.I. Comparison of proteolytic activities of the enzyme complex from mammalian pancreas and pancreatin. Appl. Biochem. Microbiol. 2000, 36, 363–367. [Google Scholar] [CrossRef]
- Andriamihaja, M.; Guillot, A.; Svendsen, A.; Hagedorn, J.; Rakotondratohanina, S.; Tomé, D.; Blachier, F. Comparative efficiency of microbial enzyme preparations versus pancreatin for in vitro alimentary protein digestion. Amino Acids 2013, 44, 563–572. [Google Scholar] [CrossRef]
- Putra, I.D.; Marsono, Y.; Indrati, R. Effect of Simulated Gastrointestinal Digestion of Bioactive Peptide from Pigeon Pea (Cajanus cajan) Tempe on Angiotensin-I Converting Enzyme Inhibitory Activity. Nutr. Food Sci. 2020, 51, 244–254. [Google Scholar] [CrossRef]
- de Castro, R.J.S.; Sato, H.H. Biologically Active Peptides: Processes for Their Generation, Purification and Identification and Applications as Natural Additives in The Food and Pharmaceutical Industries. Food Res. Int. 2015, 74, 185–198. [Google Scholar] [CrossRef]
- Zhang, C.; Wu, S.; Xu, D. Catalytic mechanism of angiotensin-converting enzyme and effects of the chloride ion. J. Phys. Chem. B 2013, 117, 6635–6645. [Google Scholar] [CrossRef]
- Pina, A.S.; Roque, A.C.A. Studies on the molecular recognition between bioactive peptides and angiotensin-converting enzyme. J. Mol. Recognit. 2009, 2008, 162–168. [Google Scholar] [CrossRef]
- Wu, S.; Feng, X.; Lan, X.; Xu, Y. Purification and identification of Angiotensin-I Converting Enzyme (ACE) inhibitory peptide from lizard fish (Saurida elongata) hydrolysate. J. Funct. Foods 2015, 13, 295–299. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hur, S.J. Antihypertensive peptides from animal products, marine organisms, and plants. Food Chem. 2017, 228, 506–517. [Google Scholar] [CrossRef]





| No | Amino Acids | Concentration (mg/L) | |
|---|---|---|---|
| <1 kDa Peptide Fraction | 1–3.5 kDa Peptide Fraction | ||
| 1 | L-arginine (R) | 102 a ± 4 | 28.1 b ± 0.3 |
| 2 | L-histidine (H) | 322 a ± 1 | 102 b ± 1 |
| 3 | L-lycine (K) | 881 a ± 15 | 331 b ± 1 |
| 4 | L-phenylalanine (F) | 86 a ± 4 | 90 a ± 1 |
| 5 | L-isoleucine (I) | 100 a ± 1 | (699.05 b ± 0.04) × 10−1 |
| 6 | L-leucine (L) | 85 b ± 2 | 120.4 a ± 0.3 |
| 7 | L-proline (P) | (0.1 ± 0.1) × 10−2 | n.d. |
| 8 | L-glycine (G) | (0.2 ± 0.1) × 10−2 | n.d. |
| 9 | L-thryptophan (W) | 0.1 a ± 0.1 | 0.02 a ± 0.02 |
| 10 | L-tyrosine (Y) | n.d. | (0.03 ± 0.02) × 10−2 |
| No | Peptide Sequence | MW (Da) | Toxicity Prediction | Activity | The Frequency of the Bioactive Fragments (A) | Potential Biological Activity of the Protein Fragments (B) | Master Protein | Accession Number |
|---|---|---|---|---|---|---|---|---|
| 1 | PRPPKPP | 787.47 | Non-toxin | ACE inhibitor | 1 | 0.04220 | OS = Glycine max | K7LI30 |
| Alpha glucosidase inhibitor | 0.28 | 1.58000 | ||||||
| DPP IV inhibitor | 0.71 | 0.00016 | ||||||
| 2 | PPPPPGARAP | 955.53 | Non-toxin | ACE inhibitor | 1.2 | 0.0056 | Formin-like protein OS = Medicago truncatula | A0A396IL77 |
| Alpha glucosidase inhibitor | 0.4 | 0.000022 | ||||||
| DPP IV inhibitor | 1.1 | 0.000125 | ||||||
| 3 | SLEGGIPR | 827.46 | Non-toxin | ACE inhibitor | 0.625 | 0.03200 | LRR receptor-like kinase resistance protein OS = Trifolium pratense | A0A2K3P9T5 |
| DPP IV inhibitor | 0.625 | 0.00035 | ||||||
| 4 | SKIVKVIGR | 998.66 | Non-toxin | ACE inhibitor | 0.3333 | 0.0087 | CTP synthase OS = Lupinus albus | A0A6A5KTD9 |
| 5 | KAGQLRK | 799.51 | Non-toxin | ACE inhibitor | 0.5714 | 0.00551 | Pentatricopeptide repeat-containing protein, mitochondrial OS = Glycine soja | A0A0B2RHL1 |
| DPP IV inhibitor | 0.5714 | 0.00002278 | ||||||
| 6 | LVNPTIPR | 908.55 | Non-toxin | ACE inhibitor | 0.5 | 0.051 | CCHC-type domain-containing protein OS = Arachis hypogaea | A0A445AQL8 |
| DPP IV inhibitor | 0.75 | 0.000304 | ||||||
| 7 | PFPPSNPPP | 948.47 | Non-toxin | ACE inhibitor | 0.8889 | 0.0035 | FAS1 domain-containing protein OS = Vigna angularis var. angularis | A0A0S3QYH1 |
| Alpha glucosidase inhibitor | 0.3333 | 0.0000184 | ||||||
| DPP IV inhibitor | 0.7778 | 0.000362 | ||||||
| 8 | IAGLDVKR | 870.53 | Non-toxin | ACE inhibitor | 0.625 | 0.01080 | OS = Lupinus albus | A0A6A5LEL5 |
| DPP IV inhibitor | 0.625 | 0.00005 | ||||||
| 9 | THGHIQVK | 918.51 | Non-toxin | ACE inhibitor | 0.375 | 0.00967 | OS = Arachis hypogaea | A0A444ZCU3 |
| DPP IV inhibitor | 0.75 | 0.00255 | ||||||
| 10 | TAPPPPPPPK | 997.57 | Non-toxin | ACE inhibitor | 1.4 | 0.03550 | OS = Trifolium subterraneum | A0A2Z6MUR4 |
| Alpha glucosidase inhibitor | 0.6 | 0.00003 | ||||||
| DPP IV inhibitor | 1.3 | 0.00012 | ||||||
| 11 | PLELVGLR | 895.56 | Non-toxin | ACE Inhibitor | 0.5 | 0.001325 | OS = Glycine soja | A0A445FEF3 |
| DPP IV inhibitor | 0.625 | 0.0000478 |
| No | Peptide Sequence | MW (Da) | Toxicity Prediction | Potential Biological Activity | The Frequency of the Bioactive Fragments (A) | Potential Biological Activity of the Protein Fragments (B) | Master Protein | Accession Number |
|---|---|---|---|---|---|---|---|---|
| 1 | VAPmSTGQATSERGA | 1477.69 | Non-toxin | ACE inhibitor | 0.57 | 0.054120 | Cellulose synthase OS = Phaseolus vulgaris | V7BSE0 |
| Dipeptidyl peptidase IV inhibitor | 0.64 | 0.000434 | ||||||
| 2 | KDGLSPDHRTLSAYID | 1786.90 | Non-toxin | ACE inhibitor | 0.38 | 0.074970 | OS = Arachis hypogaea | A0A444WW07 |
| Dipeptidyl peptidase IV inhibitor | 0.38 | 0.000034 | ||||||
| 3 | RGLPVRGQR | 1037.62 | Non-toxin | ACE inhibitor | 0.78 | 0.072000 | Adenylate kinase OS = Lupinus albus | A0A6A5LHC6 |
| Dipeptidyl peptidase IV inhibitor | 0.67 | 0.000224 | ||||||
| 4 | KATNSTAPEVNPRLLK | 1737.97 | Non-toxin | ACE inhibitor | 0.56 | 0.061000 | UV-stimulated scaffold protein A homolog isoform X1 OS = Abrus precatorius | A0A8B8L9K4 |
| alpha-glucosidase inhibitor | 0.06 | 0.000002 | ||||||
| Dipeptidyl peptidase IV inhibitor | 0.63 | 0.000018 | ||||||
| 5 | AFLPGSLVDVRPV | 1368.78 | Non-toxin | ACE inhibitor | 0.69 | 0.052600 | 30S ribosomal protein S1 OS = Lupinus albus | A0A6A5KWC6 |
| Dipeptidyl peptidase IV inhibitor | 0.77 | 0.000383 | ||||||
| 6 | GPVLEDWEKDLGPPSGG | 1751.85 | Non-toxin | ACE inhibitor | 0.65 | 0.097600 | OS = Trifolium medium | A0A392UIN2 |
| alpha-glucosidase inhibitor | 0.06 | 0.000003 | ||||||
| antioxidative | 0.12 | 0.000009 | ||||||
| Dipeptidyl peptidase IV inhibitor | 0.59 | 0.000909 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fitriani, A.; Indrati, R.; Marsono, Y.; Supriyadi, S. Impact of Gastrointestinal Digestion Simulation on the Formation of Angiotensin-I-Converting Enzyme Inhibitory (ACE-I) Peptides from Germinated Lamtoro Gung Flour. Foods 2022, 11, 3769. https://doi.org/10.3390/foods11233769
Fitriani A, Indrati R, Marsono Y, Supriyadi S. Impact of Gastrointestinal Digestion Simulation on the Formation of Angiotensin-I-Converting Enzyme Inhibitory (ACE-I) Peptides from Germinated Lamtoro Gung Flour. Foods. 2022; 11(23):3769. https://doi.org/10.3390/foods11233769
Chicago/Turabian StyleFitriani, Aprilia, Retno Indrati, Yustinus Marsono, and Supriyadi Supriyadi. 2022. "Impact of Gastrointestinal Digestion Simulation on the Formation of Angiotensin-I-Converting Enzyme Inhibitory (ACE-I) Peptides from Germinated Lamtoro Gung Flour" Foods 11, no. 23: 3769. https://doi.org/10.3390/foods11233769
APA StyleFitriani, A., Indrati, R., Marsono, Y., & Supriyadi, S. (2022). Impact of Gastrointestinal Digestion Simulation on the Formation of Angiotensin-I-Converting Enzyme Inhibitory (ACE-I) Peptides from Germinated Lamtoro Gung Flour. Foods, 11(23), 3769. https://doi.org/10.3390/foods11233769