Serotypes, Antimicrobial Resistance Profiles, and Virulence Factors of Salmonella Isolates in Chinese Edible Frogs (Hoplobatrachus rugulosus) Collected from Wet Markets in Hong Kong
Abstract
:1. Introduction
2. Materials and Methods
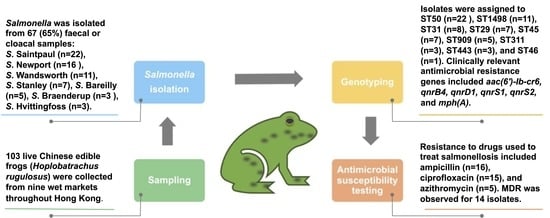
2.1. Sample Collection
2.2. Salmonella Isolation and Identification
2.3. Salmonella Serotyping
2.4. DNA Extraction and Whole Genome Sequencing
2.5. Antimicrobial Susceptibility Testing
2.6. Statistical Analysis
2.7. Ethics Statement
3. Results
3.1. Demographic Data of H. rugulosus Collected from Different Wet Markets
3.2. Prevalence of Salmonella among H. rugulosus
3.3. Serotypes
3.4. Core Genome Multilocus Sequence Types (cgMLST) and Phylogenetic Relatedness
3.5. Antimicrobial Resistance Genotypes
3.6. Antimicrobial Susceptibility Profiles
3.7. Virulence Factors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Besser, J.M. Salmonella epidemiology: A whirlwind of change. Food Microbiol. 2018, 71, 55–59. [Google Scholar] [CrossRef] [PubMed]
- Issenhuth-Jeanjean, S.; Roggentin, P.; Mikoleit, M.; Guibourdenche, M.; De Pinna, E.; Nair, S.; Fields, P.I.; Weill, F.-X. Supplement 2008–2010 (no. 48) to the White–Kauffmann–Le Minor scheme. Res. Microbiol. 2014, 165, 526–530. [Google Scholar] [CrossRef] [Green Version]
- Grimont, P.A.D.; Weill, F.-X. Antigenic formulae of the Salmonella serovars. WHO Collab. Cent. Ref. Res. Salmonella 2007, 9, 1–166. [Google Scholar]
- Achtman, M.; Wain, J.; Weill, F.-X.; Nair, S.; Zhou, Z.; Sangal, V.; Krauland, M.G.; Hale, J.L.; Harbottle, H.; Uesbeck, A.; et al. Multilocus sequence typing as a replacement for serotyping in Salmonella enterica. PLoS Pathog. 2012, 8, e1002776. [Google Scholar] [CrossRef] [Green Version]
- Chattaway, M.A.; Langridge, G.C.; Wain, J. Salmonella nomenclature in the genomic era: A time for change. Sci. Rep. 2021, 11, 7494. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2021 zoonoses report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- Ferrari, R.G.; Rosario, D.K.A.; Cunha-Neto, A.; Mano, S.B.; Figueiredo, E.E.S.; Conte-Junior, C.A. Worldwide epidemiology of Salmonella serovars in animal-based foods: A meta-analysis. Appl. Environ. Microbiol. 2019, 85, e00591-19. [Google Scholar] [CrossRef] [Green Version]
- Centers for Disease Control and Prevention. National Enteric Disease Surveillance: Salmonella Annual Report. Available online: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf (accessed on 14 April 2023).
- Li, W.; Han, H.; Liu, J.; Ke, B.; Zhan, L.; Yang, X.; Tan, D.; Yu, B.; Huo, X.; Ma, X.; et al. Antimicrobial resistance profiles of Salmonella isolates from human diarrhea cases in China: An eight-year surveillance study. One Health Adv. 2023, 1, 1–8. [Google Scholar] [CrossRef]
- Zollner-Schwetz, I.; Krause, R. Therapy of acute gastroenteritis: Role of antibiotics. Clin. Microbiol. Infect. 2015, 21, 744–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medalla, F.; Gu, W.; Mahon, B.E.; Judd, M.; Folster, J.; Griffin, P.M.; Hoekstra, R.M. Estimated incidence of antimicrobial drug-resistant nontyphoidal Salmonella infections, United States, 2004–2012. Emerg. Infect. Dis. 2016, 23, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Mølbak, K. Human health consequences of antimicrobial drug-resistant Salmonella and other foodborne pathogens. Clin. Infect. Dis. 2005, 41, 1613–1620. [Google Scholar] [CrossRef] [PubMed]
- Pulcini, C.; Beovic, B.; Béraud, G.; Carlet, J.; Cars, O.; Howard, P.; Levy-Hara, G.; Li, G.; Nathwani, D.; Roblot, F.; et al. Ensuring universal access to old antibiotics: A critical but neglected priority. Clin. Microbiol. Infect. 2017, 23, 590–592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union One Health 2019 zoonoses report. EFSA J. 2021, 19, e06406. [Google Scholar] [CrossRef]
- Altherr, S.; Auliya, M.; Nithart, C. Deadly dish—Role and Responsibility of the European Union in the International Frogs’ Legs Trade. Report by Pro Wildlife & Robin des Bois. 2022. Available online: https://www.prowildlife.de/wp-content/uploads/2022/06/DEADLY (accessed on 21 March 2023).
- Auliya, M.; Altherr, S.; Nithart, C.; Hughes, A.; Bickford, D. Numerous uncertainties in the multifaceted global trade in frogs’ legs with the EU as the major consumer. Nat. Conserv. 2023, 51, 71–135. [Google Scholar] [CrossRef]
- Grano, M. The Asian market of frogs as food for humans during COVID-19. Risk and consequences for public health. Med. Pap. 2020, 6, 77–87. [Google Scholar]
- Food and Environmental Hygiene Department. List of FEHD Public Markets/Cooked Food Markets. Available online: https://www.fehd.gov.hk/english/pleasant_environment/tidy_market/Markets_CFC_list.html (accessed on 3 May 2023).
- Underwood, W.; Raymond, A. American Veterinary Medical Association AVMA Guidelines for the Euthanasia of Animals: 2020 Edition. Available online: https://www.spandidos-publications.com/var/AVMA_euthanasia_guidelines_2020.pdf (accessed on 1 March 2023).
- Mooijman, K.A.; Pielaat, A.; Kuijpers, A.F.A. Validation of EN ISO 6579-1-Microbiology of the food chain-Horizontal method for the detection, enumeration and serotyping of Salmonella-Part 1 detection of Salmonella spp. Internat. J. Food Microbiol. 2019, 288, 3–12. [Google Scholar] [CrossRef]
- Seemann, T.S. Available online: https://githubcom/tseemann/shovill (accessed on 13 February 2023).
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic. Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Zhang, S.; Yin, Y.; Jones, M.B.; Zhang, Z.; Deatherage Kaiser, B.L.; Dinsmore, B.A.; Fitzgerald, C.; Fields, P.I.; Deng, X. Salmonella serotype determination utilizing high-throughput genome sequencing data. J. Clin. Microbiol. 2015, 53, 1685–1692. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive protein alignments at tree-of-life scale using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Liu, B.; Zheng, D.; Zhou, S.; Chen, L.; Yang, J. VFDB 2022: A general classification scheme for bacterial virulence factors. Nucleic. Acids Res. 2022, 50, D912–D917. [Google Scholar] [CrossRef] [PubMed]
- CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salipante, S.J.; Hall, B.G. Determining the limits of the evolutionary potential of an antibiotic resistance gene. Mol. Biol. Evol. 2003, 20, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Magnet, S.; Lambert, T.; Courvalin, P.; Blanchard, J.S. Kinetic and mutagenic characterization of the chromosomally encoded Salmonella enterica AAC(6’)-Iy aminoglycoside N-acetyltransferase. Biochemistry 2001, 40, 3700–3709. [Google Scholar] [CrossRef]
- Rowe-Magnus, D.A.; Mazel, D. Resistance gene capture. Curr. Opin. Microbiol. 1999, 2, 483–488. [Google Scholar] [CrossRef]
- Dos Santos, A.M.P.; Ferrari, R.G.; Conte-Junior, C.A. Virulence factors in Salmonella Typhimurium: The sagacity of a bacterium. Curr. Microbiol. 2019, 76, 762–773. [Google Scholar] [CrossRef]
- Ma, S.; Dong, Y.; Wang, N.; Liu, J.; Lu, C.; Liu, Y. Identification of a new effector-immunity pair of Aeromonas hydrophila type VI secretion system. Vet. Res. 2020, 51, 1–14. [Google Scholar] [CrossRef]
- Banerji, R.; Karkee, A.; Kanojiya, P.; Saroj, S.D. Pore-forming toxins of foodborne pathogens. Compr. Rev. Food Sci. Food Saf. 2021, 20, 2265–2285. [Google Scholar] [CrossRef]
- Auliya, M.; Altherr, S.; Hughes, A.; Nithart, C.; Ohler, A.; Bickford, D. The European market remains the largest consumer of frogs’ legs from wild species. Conservation 2023, 3, 53–58. [Google Scholar] [CrossRef]
- Ribas, A.; Poonlaphdecha, S. Wild-caught and farm-reared amphibians are important reservoirs of Salmonella, a study in North-East Thailand. Zoonoses Public Health 2017, 64, 106–110. [Google Scholar] [CrossRef]
- Prapasarakul, N.; Pulsrikarn, C.; Vasaruchapong, T.; Lekcharoen, P.; Chanchaithong, P.; Lugsomya, K.; Keschumras, N.; Thanomsuksinchai, N.; Tanchiangsai, K.; Tummaruk, P. Salmonella serovar distribution in cobras (Naja kaouthia), snake-food species, and farm workers at Queen Saovabha Snake Park, Thailand. J. Vet. Diagn. Investig. 2012, 24, 288–294. [Google Scholar] [CrossRef] [Green Version]
- Fang, Y.; Huang, H.; Li, B.; Ruan, F.; Li, Z.; Huang, W.; Wei, Q.; Huang, K. An outbreak of Salmonella Hvittingfoss infection in a tourist group back from Hong Kong, Southeast China. J. Infect. 2022, 84, e28–e30. [Google Scholar] [CrossRef] [PubMed]
- Pak, W.L.W.; Chan, K.L.; Chan, Z.; Law, W.P.; Wong, Y.H.; Lam, C.K.; Wong, S.H.S. Salmonella Wandsworth peritoneal dialysis-related peritonitis following persistent infection: A case report with a review of literature. Nephrology 2022, 27, 468–470. [Google Scholar] [CrossRef]
- Sotir, M.J.; Ewald, G.; Kimura, A.C.; Higa, J.I.; Sheth, A.; Troppy, S.; Meyer, S.; Hoekstra, R.M.; Austin, J.; Archer, J.; et al. Outbreak of Salmonella Wandsworth and Typhimurium infections in infants and toddlers traced to a commercial vegetable-coated snack food. Pediatr. Infect. Dis. J. 2009, 28, 1041–1046. [Google Scholar] [CrossRef] [PubMed]
- Aung, K.T.; Khor, W.C.; Octavia, S.; Ye, A.; Leo, J.; Chan, P.P.; Lim, G.; Wong, W.K.; Tan, B.Z.Y.; Schlundt, J. Distribution of Salmonella serovars in humans, foods, farm animals and environment, companion and wildlife animals in Singapore. Int. J. Environ. Res. Public Health 2020, 17, 5774. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Critically Important Antimicrobials for Human Medicine, 6th ed.; WHO: Geneva, Switzerland, 2019. [Google Scholar]
- Bush, K. A resurgence of β-lactamase inhibitor combinations effective against multidrug-resistant Gram-negative pathogens. Int. J. Antimicrob. Agents. 2015, 46, 483–493. [Google Scholar] [CrossRef]
- Hennequin, C.; Ravet, V.; Robin, F. Plasmids carrying DHA-1 β-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 2018, 37, 1197–1209. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Moreno, M.O.; Picó-Plana, E.; de Toro, M.; Grande-Armas, J.; Quiles-Fortuny, V.; Pons, M.J.; Gomes, C.; Sáenz, Y.; Torres, C.; Ruiz, J. β-Lactamases, transferable quinolone resistance determinants, and class 1 integron-mediated antimicrobial resistance in human clinical Salmonella enterica isolates of non-Typhimurium serotypes. Int. J. Med. Microbiol. 2013, 303, 25–31. [Google Scholar] [CrossRef]
- Robicsek, A.; Strahilevitz, J.; Sahm, D.F.; Jacoby, G.A.; Hooper, D.C. qnr prevalence in ceftazidime-resistant Enterobacteriaceae isolates from the United States. Antimicrob. Agents. Chemother. 2006, 50, 2872–2874. [Google Scholar] [CrossRef] [Green Version]
- Robicsek, A.; Strahilevitz, J.; Jacoby, G.A.; Macielag, M.; Abbanat, D.; Park, C.H.; Bush, K.; Hooper, D.C. Fluoroquinolone-modifying enzyme: A new adaptation of a common aminoglycoside acetyltransferase. Nat. Med. 2006, 12, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, K.L.; Wootton, L.; Day, M.R.; Threlfall, E.J. Plasmid-mediated quinolone resistance determinant qnrS1 found in Salmonella enterica strains isolated in the UK. J. Antimicrob. Chemother. 2007, 59, 1071–1075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.; Chen, K.; Wai-Chi Chan, E.; Chen, S. Increasing prevalence of ciprofloxacin-resistant food-borne Salmonella strains harboring multiple PMQR elements but not target gene mutations. Sci. Rep. 2015, 5, 14754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vázquez, X.; Fernández, J.; Hernáez, S.; Rodicio, R.; Rodicio, M.R. Plasmid-mediated quinolone resistance (PMQR) in two clinical strains of Salmonella enterica serovar Corvallis. Microorganisms 2022, 10, 579. [Google Scholar] [CrossRef] [PubMed]
- Chiou, C.S.; Hong, Y.P.; Wang, Y.W.; Chen, B.H.; Teng, R.H.; Song, H.Y.; Liao, Y.S. Antimicrobial resistance and mechanisms of azithromycin resistance in nontyphoidal Salmonella isolates in Taiwan, 2017 to 2018. Microbiol. Spectr. 2023, 11, e0336422. [Google Scholar] [CrossRef]
- European Food Safety Authority and European Centre for Disease Prevention and Control. The European Union summary report on antimicrobial resistance in zoonotic and indicator bacteria from humans, animals and food in 2019–2020. EFSA J. 2022, 20, 1–197. [Google Scholar] [CrossRef]
- Zhang, Z.; Yang, J.; Xu, X.; Zhou, X.; Shi, C.; Zhao, X.; Liu, Y.; Shi, X. Co-existence of mphA, oqxAB and blaCTX-M-65 on the IncHI2 plasmid in highly drug-resistant Salmonella enterica serovar Indiana ST17 isolated from retail foods and humans in China. Food Control 2020, 118, 107269. [Google Scholar] [CrossRef]
- Nguyen, Q.T.; Tran, M.P. Drugs and chemicals use in frog farming in Dong Thap province. Can. Tho. Univ. J. Sc. 2021, 13, 73–78. [Google Scholar] [CrossRef]
- Colon, V.A.; Lugsomya, K.; Lam, H.K.; Wahl, L.C.; Parkes, R.S.V.; Cormack, C.A.; Horlbog, J.A.; Stevens, M.; Stephan, R.; Magouras, I. Serotype diversity and antimicrobial resistance profile of Salmonella enterica isolates from freshwater turtles sold for human consumption in wet markets in Hong Kong. Front Vet. Sci. 2022, 9, 912693. [Google Scholar] [CrossRef]
- Hassena, A.B.; Haendiges, J.; Zormati, S.; Guermazi, S.; Gdoura, R.; Gonzalez-Escalona, N.; Siala, M. Virulence and resistance genes profiles and clonal relationships of non-typhoidal food-borne Salmonella strains isolated in Tunisia by whole genome sequencing. Intern. J. Food. Microbiol. 2021, 337, 108941. [Google Scholar] [CrossRef]

| Wet Market | Samples (n) | Positive Samples (n) | Proportion of Positive Samples (%) | Salmonella Serotypes Identified (n) |
|---|---|---|---|---|
| A | 20 | 16 | 80 | S. Newport (4), S. Saintpaul (8), S. Wandsworth (4) |
| B | 21 | 15 | 71 | S. Hvittingfoss (3), S. Newport (2), S. Saintpaul (7), S. Wandsworth (3) |
| C | 18 | 3 | 17 | S. Saintpaul (2), S. Wandsworth (1) |
| D | 6 | 6 | 100 | S. Bareilly (2), S. Stanley (4) |
| E | 10 | 10 | 100 | S. Bareilly (3), S. Newport (2), S. Saintpaul (2), S. Stanley (3) |
| F | 10 | 5 | 50 | S. Saintpaul (3), S. Wandsworth (2) |
| G | 10 | 6 | 60 | S. Newport (5), S. Wandsworth (1) |
| H | 3 | 3 | 100 | S. Braenderup (3) |
| I | 5 | 3 | 60 | S. Newport (3) |
| Isolate ID | Serotype | ST | Antimicrobial Resistance Genes a | Resistance Profile b | GenBank Accession No. |
|---|---|---|---|---|---|
| F7 | S. Bareilly | 909 | aac(6′)-Iy, ant(3″)-Ib, ant(3″)-Iia, blaTEM-1, blaTEM-60, linG, mphA, mrX, qnrS1, tet(A) | Beta-lactams (AM), macrolides (AZM), aminoglycosides (S), tetracyclines (TE) | JASBEW000000000 |
| F9 | S. Bareilly | 909 | aac(6′)-Iy, ant(3″)-Ib, ant(3″)-Iia, blaTEM-1, blaTEM-60, linG, mphA, mrX, qnrS1, tet(A) | Beta-lactams (AM), macrolides (AZM), aminoglycosides (S), tetracyclines (TE) | JASBEI000000000 |
| F61 | S. Bareilly | 909 | aac(6′)-Iy, ant(3″)-Ib, ant(3″)-Iia, blaTEM-1, blaTEM-60, linG, mphA, mrX, qnrS1, tet(A) | Beta-lactams (AM), macrolides (AZM), aminoglycosides (S), tetracyclines (TE) | JASBFI000000000 |
| F62 | S. Bareilly | 909 | aac(6′)-Iy, blaTEM-60 | – | JASBFD000000000 |
| F64 | S. Bareilly | 909 | aac(6′)-Iy, ant(3″)-Ib, ant(3″)-Iia, blaTEM-1, blaTEM-60, linG, mphA, mrX, qnrS1, tet(A) | Beta-lactams (AM), macrolides (AZM), aminoglycosides (S), tetracyclines (TE) | JASBFG000000000 |
| F1 | S. Braenderup | 311 | aac(6′)-Iaa, blaTEM-60, qnrS2 | – | JASBGW000000000 |
| F2 | S. Braenderup | 311 | aac(6′)-Iaa, blaTEM-60 | Aminoglycosides (S) | JASBGV000000000 |
| F3 | S. Braenderup | 311 | aac(6′)-Iaa, blaTEM-60, qnrS2 | Quinolones (NA) | JASBGH000000000 |
| F78 | S. Hvittingfoss | 446 | aac(6′)-Iy, blaTEM-60, qnrS2, tet(A) | Fluoroquinolones (CIP), tetracyclines (TE) | JASBET000000000 |
| F83 | S. Hvittingfoss | 446 | aac(6′)-Iy, blaTEM-60, qnrS2, tet(A) | tetracyclines (TE) | JASBEL000000000 |
| F85 | S. Hvittingfoss | 446 | aac(6′)-Iy, blaTEM-60, qnrS2, tet(A) | tetracyclines (TE) | JASBEN000000000 |
| F29 | S. Newport | 31 | aac(6′)-Iy, blaTEM-1, blaTEM-60, qnrS1, tet(A) | Beta-lactams (AM), tetracyclines (TE) | JASBGL000000000 |
| F32 | S. Newport | 31 | aac(6′)-Iy, blaTEM-1, blaTEM-60, qnrS1, tet(A) | Beta-lactams (AM), tetracyclines (TE) | JASBGC000000000 |
| F38 | S. Newport | 31 | aac(6′)-Iy, blaTEM-60 | – | JASBGA000000000 |
| F39 | S. Newport | 31 | aac(6′)-Iy, blaTEM-60, blaTEM-1, qnrS1, tet(A) | Beta-lactams (AM), tetracyclines (TE) | JASBFZ000000000 |
| F44 | S. Newport | 31 | aac(6′)-Iy, blaTEM-1, blaTEM-60, qnrS1, tet(A) | Beta-lactams (AM), tetracyclines (TE) | JASBFW000000000 |
| F55 | S. Newport | 31 | aac(6′)-Iy, blaTEM-60, dfrA14, qnrS1, tet(A) | Sulfonamides (SXT), tetracyclines (TE) | JASBFO000000000 |
| F56 | S. Newport | 31 | aac(6′)-Iy, blaTEM-60, dfrA14, qnrS1, tet(A) | Sulfonamides (SXT), tetracyclines (TE) | JASBFN000000000 |
| F67 | S. Newport | 31 | aac(6′)-Iy, blaTEM-60, dfrA14, qnrS1, tet(A) | Fluoroquinolones (CIP), sulfonamides (SXT), tetracyclines (TE) | JASBFB000000000 |
| F40 | S. Newport | 45 | aac(6′)-Ib-cr6, aac(6′)-Iy, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, arr-3, blaDHA-1, blaOXA-1, catB3, catII, qnrB4, sul1, sul2, tet(A), tet(D) | Beta-lactams (AM, AMC, CZ), amphenicols (C), fluoroquinolones (CIP), aminoglycosides (K, S), tetracyclines (TE) | JASBFX000000000 |
| F43 | S. Newport | 45 | aac(6′)-Ib-cr6, aac(6′)-Iy, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, arr-3, blaDHA-1, blaOXA-1, catB3, catII, qnrB4, sul1, sul2, tet(D) | Beta-lactams (AM, AMC, CZ), amphenicols (C), aminoglycosides (K, S), tetracyclines (TE) | JASBFV000000000 |
| F48 | S. Newport | 45 | aac(6′)-Ib-cr6, aac(6′)-Iy, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, arr-3, blaOXA-1, catB3, catII, sul1, sul2, tet(A), tet(D) | Beta-lactams (AM), amphenicols (C), fluoroquinolones (CIP), aminoglycosides (K), tetracyclines (TE) | JASBFU000000000 |
| F49 | S. Newport | 45 | aac(6′)-Ib-cr6, aac(6′)-Iy, aph(3″)-Ib, aph(6)-Id, arr-3, blaDHA-1, blaOXA-1, catB3, catII, qnrB4, sul1, sul2, tet(A), tet(D) | Beta-lactams (AM, AMC, CZ), amphenicols (C), aminoglycosides (K, S), tetracyclines (TE) | JASBFR000000000 |
| F50 | S. Newport | 45 | aac(6′)-Ib-cr6, aac(6′)-Iy, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, arr-3, blaDHA-1, blaOXA-1, catB3, catII, qnrB4, sul1, sul2, tet(A), tet(D) | Beta-lactams (AM, AMC, CZ), amphenicols (C), aminoglycosides (K, S), tetracyclines (TE) | JASBFQ000000000 |
| F51 | S. Newport | 45 | aac(6′)-Ib-cr6, aac(6′)-Iy, aph(3″)-Ib, aph(3′)-Ia, aph(6)-Id, arr-3, blaDHA-1, blaOXA-1, catB3, catII, qnrB4, sul1, sul2, tet(A), tet(D) | Beta-lactams (AM, AMC, CZ), amphenicols (C), fluoroquinolones (CIP), aminoglycosides (GM, K, S), tetracyclines (TE) | JASBFL000000000 |
| F52 | S. Newport | 45 | aac(6′)-Ib-cr6, aac(6′)-Iy, aph(3″)-Ib, aph(6)-Id, arr-3, blaDHA-1, blaOXA-1, catB3, catII, qnrB4, sul1, sul2, tet(A), tet(D) | Beta-lactams (AM, AMC, CZ), amphenicols (C), fluoroquinolones (CIP), aminoglycosides (K, S), tetracyclines (TE) | JASBFP000000000 |
| F77 | S. Newport | 46 | aac(6′)-Iy, tet(A) | Quinolones (NA), tetracyclines (TE) | JASBEQ000000000 |
| F11 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | (Fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBGS000000000 |
| F13 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGR000000000 |
| F15 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | Quinolones (NA), tetracyclines (TE) | JASBGT000000000 |
| F21 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGP000000000 |
| F23 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGO000000000 |
| F24 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGJ000000000 |
| F25 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGN000000000 |
| F26 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGI000000000 |
| F27 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGK000000000 |
| F28 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | (Fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBGM000000000 |
| F30 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBGD000000000 |
| F65 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | (Fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBFC000000000 |
| F66 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBEZ000000000 |
| F73 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | (Fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBEY000000000 |
| F75 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | (Fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBEP000000000 |
| F79 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | (Fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBEU000000000 |
| F80 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBEV000000000 |
| F92 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | (Fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBEJ000000000 |
| F59 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | Quinolones (NA), tetracyclines (TE) | JASBFE000000000 |
| F81 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBES000000000 |
| F58 | S. Saintpaul | 50 | aac(6′)-Iy, blaTEM-60 | Quinolones (NA) | JASBFK000000000 |
| F93 | S. Saintpaul | 50 | aac(6′)-Iy, ant(3″)-IIa, ant(3″)-IIa, arr-2, blaOXA-10, blaTEM-60, catI, dfrA14, dfrB4, mphA, mrX, qnrS1, sul1, sul2, tet(A) | Beta-lactams (AM), macrolides (AZM), (fluoro)quinolones (CIP, NA), tetracyclines (TE) | JASBEK000000000 |
| F4 | S. Stanley | 29 | aac(6′)-Iy, blaTEM-60 | – | JASBFT000000000 |
| F5 | S. Stanley | 29 | aac(6′)-Iy, blaTEM-60 | – | JASBFS000000000 |
| F6 | S. Stanley | 29 | aac(6′)-Iy, blaTEM-60 | – | JASBFF000000000 |
| F8 | S. Stanley | 29 | aac(6′)-Iy, blaTEM-60 | – | JASBEO000000000 |
| F60 | S. Stanley | 29 | aac(6′)-Iy, blaTEM-60 | – | JASBFH000000000 |
| F63 | S. Stanley | 29 | aac(6′)-Iy, blaTEM-60 | – | JASBFJ000000000 |
| F57 | S. Stanley | 29 | aac(6′)-Iy, blaTEM-60 | – | JASBFM000000000 |
| F12 | S. Wandsworth | 1498 | aac(6′)-Iy, blaTEM-60, qnrS2 | – | JASBGQ000000000 |
| F68 | S. Wandsworth | 1498 | aac(6′)-Iy, blaTEM-60, qnrS2 | – | JASBFA000000000 |
| F70 | S. Wandsworth | 1498 | aac(6′)-Iy, blaTEM-60, qnrS2 | – | JASBEX000000000 |
| F82 | S. Wandsworth | 1498 | aac(6′)-Ib-cr6, aac(6′)-Iy, aadA16, arr-3, blaTEM-60, dfrA27, qnrS2, sul1, tet(A) | Fluoroquinolones (CIP), sulfonamides (SXT), tetracyclines (TE) | JASBER000000000 |
| F86 | S. Wandsworth | 1498 | aac(6′)-Iy, blaTEM-60, qnrS2, vanX | – | JASBEM000000000 |
| F31 | S. Wandsworth | 1498 | aac(6′)-Ib-cr6, aac(6′)-Iy, aadA16, ant(3″)-IIa, arr-3, blaTEM-60, dfrA27, linG, qnrS1, sul1, tet(A) | Sulfonamides (SXT), tetracyclines (TE) | JASBGB000000000 |
| F33 | S. Wandsworth | 1498 | aac(6′)-Ib-cr6, aac(6′)-Iy, aadA16, ant(3″)-IIa, arr-3, blaIND-6, blaTEM-60, dfrA27, linG, qnrS1, sul1, tet(A) | Sulfonamides (SXT), tetracyclines (TE) | JASBGG000000000 |
| F35 | S. Wandsworth | 1498 | aac(6′)-Ib-cr6, aac(6′)-Iy, aadA16, ant(3″)-IIa, arr-3, blaTEM-60, dfrA27, linG, qnrS1, sul1, tet(A) | Sulfonamides (SXT), tetracyclines (TE) | JASBGE000000000 |
| F36 | S. Wandsworth | 1498 | aac(6′)-Ib-cr6, aac(6′)-Iy, aadA16, ant(3″)-IIa, arr-3, blaTEM-60, dfrA27, linG, qnrS1, sul1, tet(A) | Sulfonamides (SXT), tetracyclines (TE) | JASBGF000000000 |
| F14 | S. Wandsworth | 1498 | aac(6′)-Iy, blaTEM-60, qnrS1, tet(A) | Tetracyclines (TE) | JASBGU000000000 |
| F45 | S. Wandsworth | 1498 | aac(6′)-Iy, blaTEM-60, qnrD1 | – | JASBFY000000000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Boss, S.; Stephan, R.; Horlbog, J.A.; Magouras, I.; Colon, V.A.; Lugsomya, K.; Stevens, M.J.A.; Nüesch-Inderbinen, M. Serotypes, Antimicrobial Resistance Profiles, and Virulence Factors of Salmonella Isolates in Chinese Edible Frogs (Hoplobatrachus rugulosus) Collected from Wet Markets in Hong Kong. Foods 2023, 12, 2245. https://doi.org/10.3390/foods12112245
Boss S, Stephan R, Horlbog JA, Magouras I, Colon VA, Lugsomya K, Stevens MJA, Nüesch-Inderbinen M. Serotypes, Antimicrobial Resistance Profiles, and Virulence Factors of Salmonella Isolates in Chinese Edible Frogs (Hoplobatrachus rugulosus) Collected from Wet Markets in Hong Kong. Foods. 2023; 12(11):2245. https://doi.org/10.3390/foods12112245
Chicago/Turabian StyleBoss, Sara, Roger Stephan, Jule Anna Horlbog, Ioannis Magouras, Violaine Albane Colon, Kittitat Lugsomya, Marc J. A. Stevens, and Magdalena Nüesch-Inderbinen. 2023. "Serotypes, Antimicrobial Resistance Profiles, and Virulence Factors of Salmonella Isolates in Chinese Edible Frogs (Hoplobatrachus rugulosus) Collected from Wet Markets in Hong Kong" Foods 12, no. 11: 2245. https://doi.org/10.3390/foods12112245
APA StyleBoss, S., Stephan, R., Horlbog, J. A., Magouras, I., Colon, V. A., Lugsomya, K., Stevens, M. J. A., & Nüesch-Inderbinen, M. (2023). Serotypes, Antimicrobial Resistance Profiles, and Virulence Factors of Salmonella Isolates in Chinese Edible Frogs (Hoplobatrachus rugulosus) Collected from Wet Markets in Hong Kong. Foods, 12(11), 2245. https://doi.org/10.3390/foods12112245