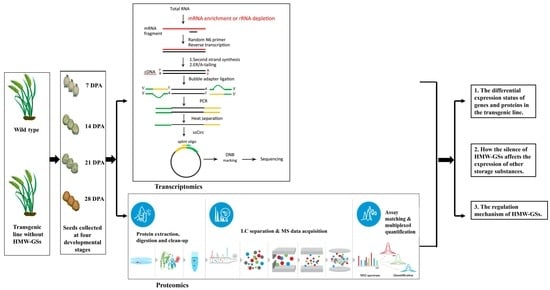
Transcriptome and Proteome Analysis Revealed the Influence of High-Molecular-Weight Glutenin Subunits (HMW-GSs) Deficiency on Expression of Storage Substances and the Potential Regulatory Mechanism of HMW-GSs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Transcriptome Analysis
2.3. DIA Proteome Analysis
2.4. Gene Expression Analyses
3. Results
3.1. Few Genes Were Commonly Identified in Transcriptome and Proteome
3.2. Accumulation Patterns of Different SSPs Varied between the Transgenic and Non-Transgenic Line
3.3. Comparison of Starch and Sucrose Metabolism-Related DEGs and DEPs
3.4. Comparisons of Amino Acids Biosynthesis-Related DEGs and DEPs
3.5. Comparisons of Transcription, Translation, and Protein Processing-Related DEGs and DEPs
4. Discussion
4.1. Absence of HMW-GSs Promotes the Accumulation of Gliadins and Certain Protease Inhibitors
4.2. Transcriptome and Proteome Studies-Related to SSPs
4.3. Coding Genes for Glutenin Genes and Transgenic Study on HMW-GS
4.4. TGS and PTGS and Their Underlying Mechanisms
4.5. Carbon and Nitrogen Metabolism
- Nitrogen
- Carbon
4.6. Translation, Transcription, and Protein Process Metabolism
4.7. Transcription Factors That Are Involved in the Regulation of the Expression of SSPs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Meziani, S.; Nadaud, I.; Tahir, A.; Nurit, E.; Benguella, R.; Branlard, G. Wheat aleurone layer: A site enriched with nutrients and bioactive molecules with potential nutritional opportunities for breeding. J. Cereal Sci. 2021, 100, 103225. [Google Scholar] [CrossRef]
- Cao, H.; He, M.; Zhu, C.; Yuan, L.; Dong, L.; Bian, Y.; Zhang, W.; Yan, Y. Distinct metabolic changes between wheat embryo and endosperm during grain development revealed by 2D-DIGE-based integrative proteome analysis. Proteomics 2016, 16, 1515–1536. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Garg, S.; Sheikh, I.; Vyas, P.; Dhaliwal, H.S. Effect of wheat grain protein composition on end-use quality. J. Food Sci. Technol. 2020, 57, 2771–2785. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Gao, J.; Zheng, X.; Feng, J.; Xue, J.; Duan, L.; Gu, Y.; Li, Y.; Yan, Y.; Li, X. Clustered Transcription Initiators and Expression of HMW-GS Genes in Wheat Endosperm. Crop Sci. 2017, 57, 378–386. [Google Scholar] [CrossRef]
- León, E.; Marín, S.; Giménez, M.J.; Piston, F.; Rodríguez-Quijano, M.; Shewry, P.R.; Barro, F. Mixing properties and dough functionality of transgenic lines of a commercial wheat cultivar expressing the 1Ax1, 1Dx5 and 1Dy10 HMW glutenin subunit genes. J. Cereal Sci. 2009, 49, 148–156. [Google Scholar] [CrossRef]
- Ma, F.; Kim, J.; Cho, E.; Brown-Guedira, G.; Park, C.S.; Baik, B.-K. HMW-GS composition and rye translocations of U.S. eastern soft winter wheat and their associations with protein strength. J. Cereal Sci. 2019, 89, 102799. [Google Scholar] [CrossRef]
- Khatkar, B.; Fido, R.; Tatham, A.; Schofield, J. Functional properties of wheat gliadins. I. Effects on mixing characteristics and bread making quality. J. Cereal Sci. 2002, 35, 299–306. [Google Scholar] [CrossRef]
- Dupont, F.M.; Vensel, W.H.; Tanaka, C.K.; Hurkman, W.J.; Altenbach, S.B. Deciphering the complexities of the wheat flour proteome using quantitative two-dimensional electrophoresis, three proteases and tandem mass spectrometry. Proteome Sci. 2011, 9, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altenbach, S.B.; Vensel, W.H.; Dupont, F.M. The spectrum of low molecular weight alpha-amylase/protease inhibitor genes expressed in the US bread wheat cultivar Butte 86. BMC Res. Notes 2011, 4, 242. [Google Scholar] [CrossRef] [Green Version]
- Barrera, G.N.; Tadini, C.C.; Leon, A.E.; Ribotta, P.D. Use of alpha-amylase and amyloglucosidase combinations to minimize the bread quality problems caused by high levels of damaged starch. J. Food Sci. Technol. 2016, 53, 3675–3684. [Google Scholar] [CrossRef]
- Ma, F.; Li, M.; Li, T.; Liu, W.; Liu, Y.; Li, Y.; Hu, W.; Zheng, Q.; Wang, Y.; Li, K.; et al. Overexpression of avenin-Like b proteins in bread wheat (Triticum aestivum L.) improves dough mixing properties by their incorporation into glutenin polymers. PLoS ONE 2013, 8, e66758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klocklewicy-Kamińska, E.; Warchalewski, J.R.; Piasecka-Kwiatkowska, D. The α-amylase, α-amylase inhibitor and proteinase inhibitor activities of proteins extracted from wheat varieties and their influence on the baking quality. Food 1995, 39, 209–218. [Google Scholar] [CrossRef]
- Pasha, I.; Anjum, F.M.; Morris, C.F. Grain hardness: A major determinant of wheat quality. Food Sci. Technol. Int. 2010, 16, 511–522. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Li, F.; Cao, S.; Zhang, K. Genomic and functional genomics analyses of gluten proteins and prospect for simultaneous improvement of end-use and health-related traits in wheat. Theor. Appl. Genet. 2020, 133, 1521–1539. [Google Scholar] [CrossRef] [Green Version]
- Juhász, A.; Belova, T.; Florides, C.G.; Maulis, C.; Fischer, I.; Gell, G.; Birinyi, Z.; Ong, J.; Keeble-Gagnère, G.; Maharajan, A.; et al. Genome mapping of seed-borne allergens and immunoresponsive proteins in wheat. Sci. Adv. 2018, 4, eaar8602. [Google Scholar] [CrossRef] [Green Version]
- Becker, D.; Wieser, H.; Koehler, P.; Folck, A.; Mühling, K.H.; Zörb, C. Protein composition and techno-functional properties of transgenic wheat with reduced α-gliadin content obtained by RNA interference. J. Appl. Bot. Food Qual. 2012, 85, 23–33. [Google Scholar]
- García-Molina, M.D.; Muccilli, V.; Saletti, R.; Foti, S.; Masci, S.; Barro, F. Comparative proteomic analysis of two transgenic low-gliadin wheat lines and non-transgenic wheat control. J. Proteom. 2017, 165, 102–112. [Google Scholar] [CrossRef]
- Marín-Sanz, M.; Iehisa, J.C.M.; Barro, F. New transcriptomic insights in two RNAi wheat lines with the gliadins strongly down-regulated by two endosperm specific promoters. Crop J. 2022, 10, 194–203. [Google Scholar] [CrossRef]
- Altenbach, S.B.; Vensel, W.H.; DuPont, F.M. Integration of transcriptomic and proteomic data from a single wheat cultivar provides new tools for understanding the roles of individual alpha gliadin proteins in flour quality and celiac disease. J. Cereal Sci. 2010, 52, 143–151. [Google Scholar] [CrossRef]
- Huo, N.; Zhu, T.; Altenbach, S.; Dong, L.; Wang, Y.; Mohr, T.; Liu, Z.; Dvorak, J.; Luo, M.-C.; Gu, Y.Q. Dynamic evolution of α-gliadin prolamin gene family in homeologous genomes of hexaploid wheat. Sci. Rep. 2018, 8, 5181. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Dai, S.; Yan, Y.; Liu, Y.; Zhang, J.; Lu, Z.; Wei, Y.; Zheng, Y.; Cong, H.; Yan, Z. The genetic diversity of group-1 homoeologs and characterization of novel LMW-GS genes from Chinese Xinjiang winter wheat landraces (Triticum aestivum L.). J. Appl. Genet. 2020, 61, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Angéla, J.; Szabolcs, M.; Endre, S.; László, T.; Ervin, B.; Battista, J.R. Role of conserved non-coding regulatory elements in LMW glutenin gene expression. PLoS ONE 2011, 6, e29501. [Google Scholar] [CrossRef] [Green Version]
- Makai, S.; Éva, C.; Tamás, L.; Juhász, A. Multiple elements controlling the expression of wheat high molecular weight glutenin paralogs. Funct. Integr. Genom. 2015, 15, 661–672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wang, K.; Li, G.; Li, Y.; Zhang, Y.; Liu, Z.; Ye, X.; Xia, X.; He, Z.; Cao, S. Dissecting conserved cis-regulatory modules of Glu-1 promoters which confer the highly active endosperm-specific expression via stable wheat transformation. Crop J. 2019, 7, 8–18. [Google Scholar] [CrossRef]
- Zhu, J.; Fang, L.; Yu, J.; Zhao, Y.; Chen, F.; Xia, G. 5-Azacytidine treatment and TaPBF-D over-expression increases glutenin accumulation within the wheat grain by hypomethylating the Glu-1 promoters. Theor. Appl. Genet. 2017, 131, 735–746. [Google Scholar] [CrossRef] [PubMed]
- Guo, D.; Hou, Q.; Zhang, R.; Lou, H.; Li, Y.; Zhang, Y.; You, M.; Xie, C.; Liang, R.; Li, B. Over-expressing TaSPA-B reduces prolamin and starch accumulation in wheat (Triticum aestivum L.) grains. Int. J. Mol. Sci. 2020, 21, 3257. [Google Scholar] [CrossRef]
- Guo, W.; Yang, H.; Liu, Y.; Gao, Y.; Ni, Z.; Peng, H.; Xin, M.; Hu, Z.; Sun, Q.; Yao, Y. The wheat transcription factor TaGAMyb recruits histone acetyltransferase and activates the expression of a high-molecular-weight glutenin subunit gene. Plant J. 2015, 84, 347–359. [Google Scholar] [CrossRef]
- Luo, G.; Shen, L.; Song, Y.; Yu, K.; Ji, J.; Zhang, C.; Yang, W.; Li, X.; Sun, J.; Zhan, K.; et al. The MYB family transcription factor TuODORANT1 from Triticum urartu and the homolog TaODORANT1 from Triticum aestivum inhibit seed storage protein synthesis in wheat. Plant Biotechnol. J. 2021, 19, 1863–1877. [Google Scholar] [CrossRef]
- Luo, G.; Shen, L.; Zhao, S.; Li, R.; Song, Y.; Song, S.; Yu, K.; Yang, W.; Li, X.; Sun, J.; et al. Genome-wide identification of seed storage protein gene regulators in wheat through coexpression analysis. Plant J. 2021, 108, 1704–1720. [Google Scholar] [CrossRef]
- Diaz, I.; Vicente-Carbajosa, J.; Abraham, Z.; Martínez, M.; Isabel-La Moneda, I.; Carbonero, P. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. Plant J. 2002, 29, 453–464. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, M.; Liu, Q.; Sun, L.; Chen, X.; Lv, L.; Liu, Y.; Jia, X.; Li, H. Deletion of high-molecular-weight glutenin subunits in wheat significantly reduced dough strength and bread-baking quality. BMC Plant Biol. 2018, 18, 319. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, L.; Zhao, A.; Zhang, Y.; Li, H.; Chen, X. Proteome and transcriptome analyses of wheat near isogenic lines identifies key proteins and genes of wheat bread quality. Sci. Rep. 2021, 11, 9978. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Choi, M.; Chang, C.-Y.; Clough, T.; Broudy, D.; Killeen, T.; MacLean, B.; Vitek, O. MSstats: An R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics 2014, 30, 2524–2526. [Google Scholar] [CrossRef] [Green Version]
- Sun, L.; Lv, L.; Zhao, J.; Hu, M.; Zhang, Y.; Zhao, Y.; Tang, X.; Wang, P.; Li, Q.; Chen, X.; et al. Genome-wide identification and expression analysis of the TaRRA gene family in wheat (Triticum aestivum L.). Front. Plant Sci. 2022, 13, 108. [Google Scholar] [CrossRef]
- Kaushik, M.; Rai, S.; Venkadesan, S.; Sinha, S.K.; Mohan, S.; Mandal, P.K. Transcriptome analysis reveals important candidate genes related to nutrient reservoir, carbohydrate metabolism, and defence proteins during grain development of hexaploid bread wheat and its diploid progenitors. Genes 2020, 11, 509. [Google Scholar] [CrossRef]
- Yang, M.; Dong, J.; Zhao, W.; Gao, X. Characterization of proteins involved in early stage of wheat grain development by iTRAQ. J. Proteom. 2016, 136, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Gao, X.; Geng, J.; Li, Q.; Li, L.; Lv, Q.; Li, X. Identification of key proteins and networks related to grain development in wheat (Triticum aestivum L.) by comparative transcription and proteomic analysis of allelic variants in TaGW2-6A. Front. Plant Sci. 2016, 7, 922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Hu, X.; Juhasz, A.; Islam, S.; Yu, Z.; Zhao, Y.; Li, G.; Ding, W.; Ma, W. Characterising avenin-like proteins (ALPs) from albumin/globulin fraction of wheat grains by RP-HPLC, SDS-PAGE, and MS/MS peptides sequencing. BMC Plant Biol. 2020, 20, 45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weber, H.; Sreenivasulu, N.; Weschke, W. Molecular physiology of seed maturation and seed storage protein biosynthesis. In Plant Developmental Biology-Biotechnological Perspectives; Springer: Berlin/Heidelberg, Germany, 2010; pp. 83–104. [Google Scholar]
- Shewry, P. What Is Gluten—Why Is It Special? Front. Nutr. 2019, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Pistón, F.; Gil-Humanes, J.; Rodríguez-Quijano, M.; Barro, F. Down-regulating γ-gliadins in bread wheat leads to non-specific increases in other gluten proteins and has no major effect on dough gluten strength. PLoS ONE 2011, 6, e24754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Altenbach, S.B.; Chang, H.-C.; Simon-Buss, A. Deciphering the immunogenic potential of wheat flour: A reference map of the salt-soluble proteome from the US wheat Butte 86. Proteome Sci. 2020, 18, 8. [Google Scholar] [CrossRef]
- Yue, S.J.; Li, H.; Li, Y.W.; Zhu, Y.F.; Guo, J.K.; Liu, Y.J.; Chen, Y.; Jia, X. Generation of transgenic wheat lines with altered expression levels of 1Dx5 high-molecular weight glutenin subunit by RNA interference. J. Cereal Sci. 2008, 47, 153–161. [Google Scholar] [CrossRef]
- Martínez-Esteso, M.J.; Nørgaard, J.; Brohée, M.; Haraszi, R.; Maquet, A.; O’Connor, G. Defining the wheat gluten peptide fingerprint via a discovery and targeted proteomics approach. J. Proteom. 2016, 147, 156–168. [Google Scholar] [CrossRef]
- Rangan, P.; Furtado, A.; Henry, R.J. The transcriptome of the developing grain: A resource for understanding seed development and the molecular control of the functional and nutritional properties of wheat. BMC Genom. 2017, 18, 766. [Google Scholar] [CrossRef] [Green Version]
- Bromilow, S.N.; Gethings, L.A.; Langridge, J.I.; Shewry, P.R.; Buckley, M.; Bromley, M.J.; Mills, E. Comprehensive proteomic profiling of wheat gluten using a combination of data-independent and data-dependent acquisition. Front. Plant Sci. 2017, 7, 2020. [Google Scholar] [CrossRef] [Green Version]
- Cho, K.; Beom, H.R.; Jang, Y.R.; Altenbach, S.B.; Vensel, W.H.; Simon-Buss, A.; Lim, S.H.; Kim, M.G.; Lee, J.Y. Proteomic Profiling and Epitope Analysis of the Complex alpha-, gamma-, and omega-Gliadin Families in a Commercial Bread Wheat. Front. Plant Sci. 2018, 9, 818. [Google Scholar] [CrossRef]
- Zhang, Y.; Lou, H.; Guo, D.; Zhang, R.; Su, M.; Hou, Z.; Zhou, H.; Liang, R.; Xie, C.; You, M. Identifying changes in the wheat kernel proteome under heat stress using iTRAQ. Crop J. 2018, 6, 600–610. [Google Scholar] [CrossRef]
- Tahir, A.; Kang, J.; Choulet, F.; Ravel, C.; Romeuf, I.; Rasouli, F.; Nosheen, A.; Branlard, G. Deciphering carbohydrate metabolism during wheat grain development via integrated transcriptome and proteome dynamics. Mol. Biol. Rep. 2020, 47, 5439–5449. [Google Scholar] [CrossRef]
- Wang, Q.; Jia, F.; Zhang, X.; Wang, X.; Li, J.; Wang, J. Transcriptome analysis reveals that the multiple metabolic pathways were related to gluten polymerization in different quality wheats (Triticum aestivum L.). Food Sci. Nutr. 2020, 8, 4573–4583. [Google Scholar] [CrossRef]
- Zimmermann, J.; Hubel, P.; Pfannstiel, J.; Afzal, M.; Longin, C.F.H.; Hitzmann, B.; Gotz, H.; Bischoff, S.C. Comprehensive proteome analysis of bread deciphering the allergenic potential of bread wheat, spelt and rye. J. Proteom. 2021, 247, 104318. [Google Scholar] [CrossRef]
- Vensel, W.H.; Tanaka, C.K.; Altenbach, S.B. Protein composition of wheat gluten polymer fractions determined by quantitative two-dimensional gel electrophoresis and tandem mass spectrometry. Proteome Sci. 2014, 12, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, N.; Chen, F.; Cui, D. Proteomic analysis of middle and late stages of bread wheat (Triticum aestivum L.) grain development. Front. Plant Sci. 2015, 6, 735. [Google Scholar] [CrossRef] [Green Version]
- Altenbach, S.B.; Chang, H.C.; Simon-Buss, A.; Mohr, T.; Huo, N.; Gu, Y.Q. Exploiting the reference genome sequence of hexaploid wheat: A proteomic study of flour proteins from the cultivar Chinese Spring. Funct. Integr. Genom. 2020, 20, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Zhen, S.; Deng, X.; Xu, X.; Liu, N.; Zhu, D.; Wang, Z.; Yan, Y. Effect of high-nitrogen fertilizer on gliadin and glutenin subproteomes during kernel development in wheat (Triticum aestivum L.). Crop J. 2020, 8, 38–52. [Google Scholar] [CrossRef]
- Dong, K.; Ge, P.; Ma, C.; Wang, K.; Yan, X.; Gao, L.; Li, X.; Liu, J.; Ma, W.; Yan, Y. Albumin and globulin dynamics during grain development of elite Chinese wheat cultivar Xiaoyan 6. J. Cereal Sci. 2012, 56, 615–622. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Li, S.; Liu, Y.; Liu, J.; Ma, X.; Du, L.; Wang, K.; Ye, X. Effects of 1Dy12 subunit silencing on seed storage protein accumulation and flour-processing quality in a common wheat somatic variation line. Food Chem. 2021, 335, 127663. [Google Scholar] [CrossRef]
- Singh, N.K.; Shepherd, K.W. Linkage mapping of genes controlling endosperm storage proteins in wheat. Theor. Appl. Genet. 1988, 75, 628–641. [Google Scholar] [CrossRef]
- Tanaka, H.; Gorafi, Y.S.A.; Fujita, M.; Sasaki, H.; Tahir, I.S.A.; Tsujimoto, H. Expression of seed storage proteins responsible for maintaining kernel traits and wheat flour quality in common wheat under heat stress conditions. Breed. Sci. 2021, 71, 184–192. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Maruyama-Funatsuki, W.; Ikeda, T.M.; Nishio, Z.; Nagasawa, K.; Tabiki, T. Dough properties and bread-making quality-related characteristics of Yumechikara near-isogenic wheat lines carrying different Glu-B3 alleles. Breed. Sci. 2015, 65, 241–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rakszegi, M.; Békés, F.; Láng, L.; Tamás, L.; Shewry, P.R.; Bedő, Z. Technological quality of transgenic wheat expressing an increased amount of a HMW glutenin subunit. J. Cereal Sci. 2005, 42, 15–23. [Google Scholar] [CrossRef]
- Vasil, I.K.; Bean, S.; Zhao, J.; McCluskey, P.; Lookhart, G.; Zhao, H.-P.; Altpeter, F.; Vasil, V. Evaluation of baking properties and gluten protein composition of field grown transgenic wheat lines expressing high molecular weight glutenin gene 1Ax1. J. Plant Physiol. 2001, 158, 521–528. [Google Scholar] [CrossRef]
- Li, S.; Liu, Y.; Tong, J.; Yu, L.; Ding, M.; Zhang, Z.; Rehman, A.U.; Majzoobi, M.; Wang, Z.; Gao, X. The overexpression of high-molecular-weight glutenin subunit Bx7 improves the dough rheological properties by altering secondary and micro-structures of wheat gluten. Food Res. Int. 2020, 130, 108914. [Google Scholar] [CrossRef]
- Roy, N.; Islam, S.; Yu, Z.; Lu, M.; Lafiandra, D.; Zhao, Y.; Anwar, M.; Mayer, J.E.; Ma, W. Introgression of an expressed HMW 1Ay glutenin subunit allele into bread wheat cv. Lincoln increases grain protein content and breadmaking quality without yield penalty. Theor. Appl. Genet. 2020, 133, 517–528. [Google Scholar] [CrossRef]
- Garg, M.; Tanaka, H.; Ishikawa, N.; Takata, K.; Yanaka, M.; Tsujimoto, H. Agropyron elongatum HMW-glutenins have a potential to improve wheat end-product quality through targeted chromosome introgression. J. Cereal Sci. 2009, 50, 358–363. [Google Scholar] [CrossRef]
- Gao, X.; Liu, T.; Ding, M.; Wang, J.; Li, C.; Wang, Z.; Li, X. Effects of HMW-GS Ax1 or Dx2 absence on the glutenin polymerization and gluten micro structure of wheat (Triticum aestivum L.). Food Chem. 2018, 240, 626–633. [Google Scholar] [CrossRef]
- Sanders, M.; Maddelein, W.; Depicker, A.; Van Montagu, M.; Cornelissen, M.; Jacobs, J. An active role for endogenous beta-1,3-glucanase genes in transgene-mediated co-suppression in tobacco. EMBO J. 2002, 21, 5824–5832. [Google Scholar] [CrossRef]
- Vaucheret, H.; Béclin, C.; Elmayan, T.; Feuerbach, F.; Godon, C.; Morel, J.-B.; Mourrain, P.; Palauqui, J.-C.; Vernhettes, S. Transgene-induced gene silencing in plants. Plant J. 1998, 16, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, H.; Hirochika, H. Silencing of transposable elements in plants. Trends Plant Sci. 2001, 6, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Ohno, S.; Makishima, R.; Doi, M. Post-transcriptional gene silencing of CYP76AD controls betalain biosynthesis in bracts of bougainvillea. J. Exp. Bot. 2021, 72, 6949–6962. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhu, Z.; Du, P.; Zhang, C.; Yan, H.; Wang, W.; Li, W. NtRBP45, a nuclear RNA-binding protein of Nicotiana tabacum, facilitates post-transcriptional gene silencing. Plant Direct 2020, 4, e00294. [Google Scholar] [CrossRef] [PubMed]
- Iyer, L.M.; Kumpatla, S.P.; Chandrasekharan, M.B.; Hall, T.C. Transgene silencing in monocots. Plant Mol. Biol. 2000, 43, 323–346. [Google Scholar] [CrossRef]
- Hollister, J.D.; Gaut, B.S. Epigenetic silencing of transposable elements: A trade-off between reduced transposition and deleterious effects on neighboring gene expression. Genome Res. 2009, 19, 1419–1428. [Google Scholar] [CrossRef] [Green Version]
- Jordan, K.W.; He, F.; de Soto, M.F.; Akhunova, A.; Akhunov, E. Differential chromatin accessibility landscape reveals structural and functional features of the allopolyploid wheat chromosomes. Genome Biol. 2020, 21, 176. [Google Scholar] [CrossRef]
- Butel, N.; Yu, A.; Le Masson, I.; Borges, F.; Elmayan, T.; Taochy, C.; Gursanscky, N.R.; Cao, J.; Bi, S.; Sawyer, A. Contrasting epigenetic control of transgenes and endogenous genes promotes post-transcriptional transgene silencing in Arabidopsis. Nat. Commun. 2021, 12, 2787. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, M.; Wang, G.; Galili, G. New insights into the metabolism of aspartate-family amino acids in plant seeds. Plant Reprod. 2018, 31, 203–211. [Google Scholar] [CrossRef]
- Altenbach, S.B. Proteomics of wheat flour. In Proteomics in Food Science; Elsevier: Amsterdam, The Netherlands, 2017; pp. 57–73. [Google Scholar]
- Song, L.; Li, L.; Zhao, L.; Liu, Z.; Xie, T.; Li, X. Absence of Dx2 at Glu-D1 locus weakens gluten quality potentially regulated by expression of nitrogen metabolism enzymes and glutenin-related genes in wheat. Int. J. Mol. Sci. 2020, 21, 1383. [Google Scholar] [CrossRef] [PubMed]
- Weber, H.; Borisjuk, L.; Wobus, U. Sugar import and metabolism during seed development. Trends Plant Sci. 1997, 2, 169–174. [Google Scholar] [CrossRef]
- Weichert, N.; Saalbach, I.; Weichert, H.; Kohl, S.; Erban, A.; Kopka, J.; Hause, B.; Varshney, A.; Sreenivasulu, N.; Strickert, M.; et al. Increasing sucrose uptake capacity of wheat grains stimulates storage protein synthesis. Plant Physiol. 2010, 152, 698–710. [Google Scholar] [CrossRef] [Green Version]
- Jeon, J.-S.; Ryoo, N.; Hahn, T.-R.; Walia, H.; Nakamura, Y. Starch biosynthesis in cereal endosperm. Plant Physiol. Biochem. 2010, 48, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Mukherjee, S.; Ayele, B.T. Molecular aspects of sucrose transport and its metabolism to starch during seed development in wheat: A comprehensive review. Biotechnol. Adv. 2018, 36, 954–967. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, W.; Qi, J.; Shi, P.; Yin, Y. Starch accumulation, activities of key enzyme and gene expression in starch synthesis of wheat endosperm with different starch contents. J. Food Sci. Technol. 2014, 51, 419–429. [Google Scholar] [CrossRef] [Green Version]
- Vinje, M.A.; Willis, D.K.; Duke, S.H.; Henson, C.A. Differential expression of two β-amylase genes (Bmy1 and Bmy2) in developing and mature barley grain. Planta 2011, 233, 1001–1010. [Google Scholar] [CrossRef]
- Xue, D.; Dong, J.; Wu, F.; Zhang, G. Genotypic and environmental variation in grain protein components and their relations to beta-amylase and beta-glucanase activity in malting barley. Cereal Res. Commun. 2008, 36, 125–134. [Google Scholar] [CrossRef]
- Call, L.; Haider, E.; D’Amico, S.; Reiter, E.; Grausgruber, H. Synthesis and accumulation of amylase-trypsin inhibitors and changes in carbohydrate profile during grain development of bread wheat (Triticum aestivum L.). BMC Plant Biol. 2021, 21, 113. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Takaiwa, F. Cereal seed storage protein synthesis: Fundamental processes for recombinant protein production in cereal grains. Plant Biotechnol. J. 2010, 8, 939–953. [Google Scholar] [CrossRef]
- Hu, S.-F.; Wei, W.-L.; Hong, S.-F.; Fang, R.-Y.; Wu, H.-Y.; Lin, P.-C.; Sanobar, N.; Wang, H.-P.; Sulistio, M.; Wu, C.-T.; et al. Investigation of the effects of P1 on HC-pro-mediated gene silencing suppression through genetics and omics approaches. Bot. Stud. 2020, 61, 22. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Li, Y.; Ma, Z.; Yan, X.; Li, N.; Shang, B.; Hu, X.; Cui, K.; Koiwa, H.; Zhang, X. The epigenetic factor FVE orchestrates cytoplasmic SGS3-DRB4-DCL4 activities to promote transgene silencing in Arabidopsis. Sci. Adv. 2021, 7, eabf3898. [Google Scholar] [CrossRef]
- Zhang, S.; Ghatak, A.; Bazargani, M.M.; Bajaj, P.; Varshney, R.K.; Chaturvedi, P.; Jiang, D.; Weckwerth, W. Spatial distribution of proteins and metabolites in developing wheat grain and their differential regulatory response during the grain filling process. Plant J. 2021, 107, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Li, H.; Wang, K.; Yang, J.; Duan, M.; Zhang, J.; Ye, N. Regulation of gene expression involved in the remobilization of rice straw carbon reserves results from moderate soil drying during grain filling. Plant J. 2020, 101, 604–618. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, V.; Parihar, V.; Malik, G.; Kalra, V.; Kapoor, S.; Kapoor, M. The DEAD-box RNA helicase eIF4A regulates plant development and interacts with the hnRNP LIF2L1 in Physcomitrella patens. Mol. Genet. Genom. 2020, 295, 373–389. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, V.; Parihar, V.; Singh, D.; Kapoor, S.; Kapoor, M. The DEAD-box RNA helicase eIF4A1 interacts with the SWI2/SNF2-related chromatin remodelling ATPase DDM1 in the moss Physcomitrella. Biochim. Biophys. Acta (BBA) Proteins Proteom. 2021, 1869, 140592. [Google Scholar] [CrossRef]
- Chantha, S.-C.; Emerald, B.S.; Matton, D.P. Characterization of the plant Notchless homolog, a WD repeat protein involved in seed development. Plant Mol. Biol. 2006, 62, 897–912. [Google Scholar] [CrossRef]
- Bonnot, T.; Martre, P.; Hatte, V.; Dardevet, M.; Leroy, P.; Bénard, C.; Falagán, N.; Martin-Magniette, M.-L.; Deborde, C.; Moing, A.; et al. Omics data reveal putative regulators of einkorn grain protein composition under sulfur deficiency. Plant Physiol. 2020, 183, 501–516. [Google Scholar] [CrossRef] [Green Version]
- Zheng, T.; Qi, P.-F.; Cao, Y.-L.; Han, Y.-N.; Ma, H.-L.; Guo, Z.-R.; Wang, Y.; Qiao, Y.-Y.; Hua, S.-Y.; Yu, H.-Y.; et al. Mechanisms of wheat (Triticum aestivum) grain storage proteins in response to nitrogen application and its impacts on processing quality. Sci. Rep. 2018, 8, 11928. [Google Scholar] [CrossRef] [Green Version]
- Ding, H.; Mo, S.; Qian, Y.; Yuan, G.; Wu, X.; Ge, C. Integrated proteome and transcriptome analyses revealed key factors involved in tomato (Solanum lycopersicum) under high temperature stress. Food Energy Secur. 2020, 9, e239. [Google Scholar] [CrossRef]
- Yu, X.; Wang, L.; Ran, L.; Chen, X.; Sheng, J.; Yang, Y.; Wu, Y.; Chen, G.; Xiong, F. New insights into the mechanism of storage protein biosynthesis in wheat caryopsis under different nitrogen levels. Protoplasma 2020, 257, 1289–1308. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; An, K.; Guo, W.; Chen, Y.; Zhang, R.; Zhang, X.; Chang, S.; Rossi, V.; Jin, F.; Cao, X. The endosperm-specific transcription factor TaNAC019 regulates glutenin and starch accumulation and its elite allele improves wheat grain quality. Plant Cell 2021, 33, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Xie, L.; Tian, X.; Liu, S.; Xu, D.; Jin, H.; Song, J.; Dong, Y.; Zhao, D.; Li, G.; et al. TaNAC100 acts as an integrator of seed protein and starch synthesis exerting pleiotropic effects on agronomic traits in wheat. Plant J. 2021, 108, 829–840. [Google Scholar] [CrossRef] [PubMed]





| Analysis | Developmental Stage | Upregulated | Downregulated | Total |
|---|---|---|---|---|
| Transcriptome | 7 DPA | 113 | 124 | 237 |
| 14 DPA | 108 | 90 | 198 | |
| 21 DPA | 737 | 854 | 1591 | |
| 28 DPA | 1088 | 256 | 1344 | |
| Proteome | 7 DPA | 46 | 186 | 232 |
| 14 DPA | 70 | 40 | 110 | |
| 21 DPA | 64 | 80 | 144 | |
| 28 DPA | 58 | 170 | 228 |
| Stage | Gene ID | Gene Description |
|---|---|---|
| 7 DPA | TraesCS4A02G036200 | 4-coumarate-CoA ligase family protein |
| TraesCS2B02G292100 | Aldo/keto reductase family protein | |
| TraesCS3B02G102100 | Fatty acid oxidation complex subunit alpha | |
| 14 DPA | TraesCS4A02G036200 | 4-coumarate-CoA ligase family protein |
| TraesCS7B02G038200 | Chalcone-flavonone isomerase | |
| TraesCSU02G117700 | DnaJ domain protein | |
| TraesCS2A02G566600 | DDB1- and CUL4-associated factor homolog 1 | |
| 21 DPA | TraesCS3B02G584700 | Pathogenesis-related protein PR-4 |
| TraesCS7B02G038200 | Chalcone-flavonone isomerase | |
| TraesCS4A02G130800 | Ricin B-like lectin R40C1 | |
| TraesCS7A02G036000 | Transmembrane protein, putative | |
| TraesCS7D02G535500 | 1,4-alpha-glucan-branching enzyme | |
| TraesCSU02G117700 | DnaJ domain protein | |
| TraesCS5D02G004000 | Grain softness protein | |
| 28 DPA | TraesCS4A02G453600 | Gliadin-like avenin |
| TraesCS2B02G392500 | Kunitz trypsin inhibitor | |
| TraesCS4B02G173700 | Ricin B-like lectin R40C1 | |
| TraesCS4A02G130800 | Ricin B-like lectin R40C1 | |
| TraesCS2B02G292100 | Probable aldo-keto reductase 2 | |
| TraesCS4A02G092900 | Heat-shock protein | |
| TraesCSU02G117700 | DnaJ domain protein | |
| TraesCS1A02G233200 | Signal peptidase I | |
| TraesCS7A02G070900 | Peroxidase | |
| TraesCS5A02G424800 | Dehydrin | |
| TraesCS1B02G322900 | Transcription elongation factor SPT6 | |
| TraesCS7A02G325700 | Seed maturation protein |
| 7 DPA | 14 DPA | 21 DPA | 28 DPA | |||||
|---|---|---|---|---|---|---|---|---|
| Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | Upregulated | Downregulated | |
| Gliadins | 0 | 0 | 1 | 0 | 30 | 0 | 5 | 2 |
| ALPs | 0 | 0 | 3 | 0 | 1 | 10 | 0 | 6 |
| Globlins | 0 | 0 | 0 | 0 | 18 | 0 | 5 | 3 |
| HMW-GSs and LMW-GSs | 0 | 0 | 0 | 6 | 4 | 6 | 0 | 7 |
| nsLTPs | 0 | 6 | 0 | 1 | 13 | 5 | 1 | 0 |
| Protease inhibitors | 0 | 0 | 0 | 0 | 19 | 11 | 18 | 3 |
| Stage | Gene ID | Fold Change | Description |
|---|---|---|---|
| 7 DPA | TraesCS1B02G059000 | −1.21527 | 11S globulin seed storage protein 2 |
| TraesCS2A02G375400 | −1.37033 | Kunitz trypsin inhibitor | |
| TraesCS2B02G392500 | −2.65509 | Kunitz trypsin inhibitor | |
| TraesCS2D02G371800 | −1.13515 | Kunitz trypsin inhibitor | |
| TraesCS3D02G323200 | −1.12579 | Protease inhibitor/seed storage/lipid transfer protein family protein | |
| 14 DPA | TraesCS1A02G007700 | −1.10619 | Gamma gliadin |
| TraesCS5A02G432100 | 1.535714 | Globulin-1 | |
| TraesCS4A02G135500 | 1.444786 | Vicilin-like seed protein | |
| 21 DPA | TraesCS2B02G471000 | 1.595181 | Protease inhibitor/seed storage/lipid transfer family protein |
| TraesCS2D02G449000 | 1.386938 | Protease inhibitor/seed storage/lipid transfer family protein | |
| TraesCS2D02G449100 | 1.127106 | Protease inhibitor/seed storage/lipid transfer family protein | |
| TraesCS5A02G424800 | −1.52425 | Dehydrin | |
| TraesCS5B02G426700 | −1.26968 | Dehydrin | |
| TraesCS5B02G426800 | −1.21027 | Dehydrin | |
| TraesCS5D02G004000 | −1.25508 | Grain softness protein | |
| TraesCSU02G108500 | −1.72641 | Alpha-gliadin | |
| TraesCS4B02G393400 | −1.16845 | Lipid-transfer protein | |
| 28 DPA | TraesCS1D02G046400 | −1.22112 | 11S globulin seed storage protein |
| TraesCS3B02G062600 | −1.92171 | Non-specific lipid-transfer protein | |
| TraesCS5B02G145900 | −1.28086 | Non-specific lipid-transfer protein | |
| TraesCS5D02G004000 | −1.34982 | Grain softness protein | |
| TraesCS7D02G504800 | −1.14673 | Puroindoline b | |
| TraesCS2B02G392500 | −1.23746 | Kunitz trypsin inhibitor | |
| TraesCS3B02G515100 | −1.37625 | Basic 7S globulin | |
| TraesCS4A02G453600 | −2.67785 | Gliadin-like avenin | |
| TraesCS5D02G145300 | −1.09181 | Non-specific lipid-transfer protein | |
| TraesCS5A02G424800 | −1.11499 | Dehydrin |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, Y.; Zhao, J.; Hu, M.; Sun, L.; Liu, Q.; Zhang, Y.; Li, Q.; Wang, P.; Ma, W.; Li, H.; et al. Transcriptome and Proteome Analysis Revealed the Influence of High-Molecular-Weight Glutenin Subunits (HMW-GSs) Deficiency on Expression of Storage Substances and the Potential Regulatory Mechanism of HMW-GSs. Foods 2023, 12, 361. https://doi.org/10.3390/foods12020361
Zhao Y, Zhao J, Hu M, Sun L, Liu Q, Zhang Y, Li Q, Wang P, Ma W, Li H, et al. Transcriptome and Proteome Analysis Revealed the Influence of High-Molecular-Weight Glutenin Subunits (HMW-GSs) Deficiency on Expression of Storage Substances and the Potential Regulatory Mechanism of HMW-GSs. Foods. 2023; 12(2):361. https://doi.org/10.3390/foods12020361
Chicago/Turabian StyleZhao, Yun, Jie Zhao, Mengyun Hu, Lijing Sun, Qian Liu, Yelun Zhang, Qianying Li, Peinan Wang, Wujun Ma, Hui Li, and et al. 2023. "Transcriptome and Proteome Analysis Revealed the Influence of High-Molecular-Weight Glutenin Subunits (HMW-GSs) Deficiency on Expression of Storage Substances and the Potential Regulatory Mechanism of HMW-GSs" Foods 12, no. 2: 361. https://doi.org/10.3390/foods12020361
APA StyleZhao, Y., Zhao, J., Hu, M., Sun, L., Liu, Q., Zhang, Y., Li, Q., Wang, P., Ma, W., Li, H., Gao, H., & Zhang, Y. (2023). Transcriptome and Proteome Analysis Revealed the Influence of High-Molecular-Weight Glutenin Subunits (HMW-GSs) Deficiency on Expression of Storage Substances and the Potential Regulatory Mechanism of HMW-GSs. Foods, 12(2), 361. https://doi.org/10.3390/foods12020361