A Role of Multi-Omics Technologies in Sheep and Goat Meats: Progress and Way Ahead
Abstract
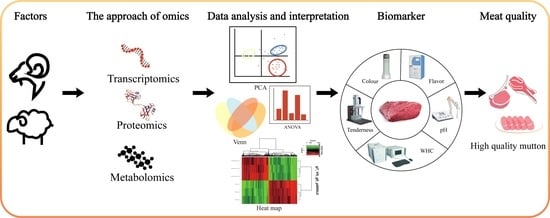
:1. Introduction
2. Transcriptomics
2.1. Overview of Transcriptomics
2.2. Transcriptomics Techniques
2.3. Application of Transcriptomics in Sheep and Goat Meat Research
2.3.1. Differences in IMF Deposition across Diverse Breeds
2.3.2. Effects of Different Feeding Managements on IMF Deposition
| Breed (Species) | Technology | Muscle/ Tissue | Treatments | Factors | Meat Traits | Ref. |
|---|---|---|---|---|---|---|
| Barbari and Changthangi goats (Capra hircus) | RNA-Seq (Illumina) | Longissimus thoracis | Four Barbari goats and four Changthangi goats with the same growing environment and feeding management were selected. | Breed | IMF content | [1] |
| Dorper × Small-tailed Han sheep (Ovis aries) | RNA-Seq (Illumina) | Longissimus dorsi | Fifty Dorper × Small-tailed Han and fifty Small-tailed Han sheep were fed 3 times a day (6 a.m., 12 p.m., 18 p.m.) in the same environment for 6 months. | Breed | Color and tenderness | [30] |
| Dorper and Small-tailed Han sheep (Ovis aries) | RNA-Seq (Illumina) | Longissimus dorsi | Dorper and Small-tailed Han sheep with similar age and weight were selected and fed for 6 months under the same feeding management. | Breed | Tenderness | [31] |
| Dorper sheep (Ovis aries) | RNA-Seq (Illumina) | Longissimus dorsi | Twelve Dorper × Hu crossbred weaning male sheep were fed with different proportions of fermented diets. The mixed proportions of Broussonetia papyrifera L treated with fermented feed were 0%, 6%, 18%, and 100% of the total diet. | Feeding manage-ment | Tenderness | [32] |
| Sunit sheep (Capra hircus) | RNA-Seq (Illumina) | Longissimus thoracis | Twelve 3-month-old Sunit lambs were divided into two groups. The experimental group was supplemented with 1% probiotic feed and fed for 90 days. | Feeding manage-ment | / | [35] |
| Small-tailed Han sheep (Ovis aries) | RNA-Seq (Illumina) | Left hepatic lobe | Thirty 3-month-old Small-tailed Han sheep were randomly divided into two groups. The experimental group was supplemented with allium mongolicum extract. After 75 days of feeding, 12 sheep in each group were randomly selected for slaughter. | Feeding manage-ment | Flavor | [36] |
| Sunit sheep (Capra hircus) | RT-PCR | Longissimus dorsi | Twenty-four Sunit sheep with the same genetic background were randomly divided into two groups, namely free grazing group and house feeding group. | Feeding manage-ment | Color | [37] |
| Tibetan sheep (Ovis aries) | RNA-Seq (Illumina) | Longissimus thoracis | The meat samples from Tibetan sheep aged 4 months, 1.5, 3.5, and 6 years were collected from the same feeding environment. | Growth stages | Tenderness | [38] |
3. Proteomics
3.1. Overview of Proteomics
3.2. Proteomics Techniques
3.3. Application of Proteomics in Sheep and Goat Meat Research
3.3.1. Changes in Meat Quality during Storage
3.3.2. Effects of Slaughtering Methods on Meat Quality
3.3.3. Effects of Processing Methods on Meat Quality
3.3.4. Effects of Post-Mortem Protein Modification on Meat Quality
| Breed (Species) | Technology | Muscle | Treatments | Factors | Meat Traits | Ref. |
|---|---|---|---|---|---|---|
| Capra hircus | iTRAQ | Longissimus thoracis | The meat samples were collected after 0, 1, and 2 freeze–thaw cycles, respectively. | Freeze–thaw cycles | Color and tenderness | [42] |
| Hengshan goats (Capra hircus) | DIA | Longissimus lumborum | After frozen for 0, 30, and 60 days, the meat samples were collected for proteomics analysis. | Freezing times | Color and tenderness | [43] |
| Nellore sheep (Ovis aries) | 2-DE/MS | Longissimus thoracis et lumborum | Fifteen Nellore sheep were slaughtered without prior electrical stunning, and the other sheep were electrically stunned. | Slaughtering method | Ultimate pH and tenderness | [44] |
| Hengshan goats (Capra hircus) | DIA | Longissimus | The meat samples were collected after boiling, roasting, and steaming processing. | Thermal processing | Tenderness | [46] |
| Fat-tailed sheep × Small-tailed Han sheep (Ovis aries) | 2-DE/MS | Longissimus thoracis et lumborum | Forty 6-month-old, uncastrated sheep were collected at 0.5, 4, 12, and 24 h post mortem. | Storage time | Tenderness | [48] |
4. Metabolomics
4.1. Overview of Metabolomics
4.2. Metabolomics Techniques
4.3. Application of Metabolomics in Sheep and Goat Meat Research
4.3.1. Strategies for Improving Meat Quality Based on Metabolomics
4.3.2. Applications of Lipidomics and Flavor Metabolomics in Improving Meat Quality
| Breed (Species) | Technology | Muscle | Treatments | Factors | Meat Traits | Ref. |
|---|---|---|---|---|---|---|
| Lubei White, Jining Gray, and Boer goats (Capra hircus) | LC-MS | Latissimus dorsi | Minimally invasive muscle biopsy under local anesthesia in 6-month-old goats of three different breeds was performed. | Breed | Flavor | [54] |
| Tan sheep (Ovis aries) | LC-MS | Lumborum | Twenty-four Tan sheep aged 120 days were randomly divided into three groups: (1) indoor feeding (F); (2) artificial pasture grazing with indoor feeding (GF); (3) pure artificial pasture grazing (G). | Feeding management | Flavor | [3] |
| Tan sheep (Ovis aries) | GC-MS | Hind legs | The surfaces of refrigerated sheep hind legs were scraped on day 0, 4, and 8 in order to collect surface microbes, blood, and exudate. | Chilled storage | Freshness level | [56] |
| Ujimqin sheep, Sunit sheep, Small-tailed Han sheep, Boer goats, and cashmere goats (Ovis aries/Capra hircus) | UPLC-Q-TOF/MS | Biceps femoris | A total of 138 sheep/goats meat samples of two types (pasture-fed and concentrate-fed) were collected. | Feeding management | Color and flavor | [58] |
| Hu sheep (Ovis aries) | UHPLC-Q-Orbitrap MS and SPME-GC-MS | Longissimus lumborum | The meat samples were collected from male Hu sheep, including 12 samples with high IMF content, and 12 samples with low IMF content. | Individuality | Flavor and IMF | [59] |
| Hu sheep (Ovis aries) | UPLC Q-Exactive Orbitrap MS | Psoas major | Twenty-four Hu sheep with the same genetic background were divided into a castration group and a control group. The sheep were raised under the same environmental conditions for 27 weeks. | Feeding management | Flavor | [60] |
| Tan sheep (Ovis aries) | HS-SPME-GC-MS/GC-MS | Longissimus dorsi | The meat samples from 10 Tan sheep were packaged in plastic film and stored at 4 ± 1 °C for 0, 1, 3, 5, and 7 d. | Storage time | Flavor | [61] |
| Mongolian sheep (Ovis aries) | UPLC-ESI-MS/MS | Longissimus thoracis | The meat samples were collected from six Mongolian ewes and stored at 4 °C for 0, 24, 48, 72, and 96 h. | Storage time | Flavor | [62] |
| Tan sheep (Ovis aries) | UPLC-ESI-MS | Longissimus dorsi | The meat samples from ten 4-year-old male Tan sheep were collected, blended, and stored in PE plastic bags for up to 24 days at −20 °C. | Storing time | Flavor and eating quality | [63] |
| Tan sheep/Hengshan goats (Ovis aries/Capra hircus) | UHPLC-Q-Orbitrap MS/MS | Longissimus thoracis et lumborum/Longissimus dorsi | The meat samples were collected from 10 four-year-old Tan sheep and treated with different concentrations of nisin and potassium sorbate preservatives. | Preservatives | Lipid composition | [64] |
| Tan sheep (Ovis aries) | UHPLC-Q-Orbitrap HRMS | Longissimus dorsi | The meat samples were cooked by different methods for set times and temperatures. | Thermal processing | Lipid composition | [66] |
| Small-tailed sheep × Mongolian sheep (Ovis aries) | UPLC-ESI-MS/MS and Orbitrap Exploris GC | back strap | The meat samples were roasted for 0, 2.5, 5, 7.5, 10, and 15 min using the traditional charcoal method. | Thermal processing | Lipid composition | [67] |
5. Multi-Omics
5.1. Overview of Multi-Omics
5.2. Integrated Transcriptomics and Proteomics
5.3. Integrated Proteomics and Metabolomics
5.4. Integrated Transcriptomics and Metabolomics
6. Conclusive Remarks and Future Perspective
6.1. Conclusive Remarks
6.2. Future Perspectives in Application of Omics Technology
- i.
- In applying omics to sheep and goat meat quality research, stable isotope tracing technology can be used to reflect the changes in intracellular substances. This would be more conducive to revealing the relevant mechanisms regulating meat quality. However, there is currently a lack of studies employing this method.
- ii.
- As a robust tool for high-throughput screening, multi-omics research lacks reasonable verification when used for data annotation and enrichment.
- iii.
- When multi-omics strategies are used to search for biomarkers, it is often difficult to find biomarkers for a single meat quality trait. The callipyge gene promotes muscle growth, but inhibits fat deposition and reduces tenderness. Additionally, heat shock proteins have an inhibitory effect on tenderness, while maintaining color stability.
- iv.
- The accuracy and preference of multi-omics technologies somewhat affect their authenticity. Currently, after optimizing the data acquisition methodology, detection coverage was enhanced during experimental validation, though some transcripts, proteins, and metabolites with lower expression abundances remained undetected, resulting in a lack of complete data in the multi-omics databases.
- v.
- Efficient and scientific methods are still needed for correlating and integrating the datasets from multidimensional omics studies of transcriptomes, proteomes, and metabolomes.
- vi.
- There is an insufficient crossover between differentiated disciplines. It is necessary to perform differentiation and synthesis to ensure high-quality meat production.
6.3. Directions for Addressing These Challenges
- i.
- Strengthening the application of tracer technology is crucial for studying sheep and goat meat quality and elucidating the regulatory mechanisms. Attention should be paid to intracellular substance dynamics to better explain the mechanisms related to meat quality.
- ii.
- When multi-omics technologies are used to explore meat quality mechanisms, potential molecular biomarkers require validation in a reasonable manner.
- iii.
- The exploration of biomarkers for individual meat quality attributes should be emphasized to facilitate the potential targeted control of meat quality.
- iv.
- High-throughput multi-omic technologies can verify each other. State-of-the-art omics technologies, including third-generation transcriptomics, ‘4D’ proteomics, and spatial omics, enable rapid identification, deep coverage, and high accuracy. Applying these advanced techniques in future meat quality studies is recommended.
- v.
- A multi-omics database with genes as connection points should be established, along with integration algorithms and software for multi-omics data.
- vi.
- Utilizing interdisciplinary knowledge in research on sheep/goat biomarkers related to meat quality can improve breeding evaluation accuracy, reduce the associated costs, and accelerate the breeding process. Combining this with proper feeding, slaughter, transportation, and processing management has great potential to produce high-quality meat.
Author Contributions
Funding
Conflicts of Interest
References
- Kumar, A.; Kaur, M.; Ahlawat, S.; Sharma, U.; Singh, M.K.; Singh, K.V.; Chhabra, P.; Vijh, R.K.; Yadav, A.; Arora, R. Transcriptomic Diversity in Longissimus Thoracis Muscles of Barbari and Changthangi Goat Breeds of India. Genomics 2021, 113, 1639–1646. [Google Scholar] [CrossRef]
- Wei, Y.C.; Li, X.; Zhang, D.Q.; Liu, Y.F. Comparison of Protein Differences between High- and Low-Quality Goat and Bovine Parts Based on iTRAQ Technology. Food Chem. 2019, 289, 240–249. [Google Scholar] [CrossRef]
- Wang, B.; Wang, Y.J.; Zuo, S.X.; Peng, S.J.; Wang, Z.J.; Zhang, Y.J.; Luo, H.L. Untargeted and Targeted Metabolomics Profiling of Muscle Reveals Enhanced Meat Quality in Artificial Pasture Grazing Tan Lambs via Rescheduling the Rumen Bacterial Community. J. Agric. Food Chem. 2021, 69, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, N.T. Sheep Welfare During Transport and Slaughter in Bulgaria—Impact of Welfare on Slaughter Carcass and Meat Quality: A Review. Turk. J. Vet. Anim. Sci. 2020, 44, 174–181. [Google Scholar] [CrossRef]
- Prache, S.; Schreurs, N.; Guillier, L. Review: Factors Affecting Sheep Carcass and Meat Quality Attributes. Animal 2022, 16, 100330. [Google Scholar] [CrossRef] [PubMed]
- Gagaoua, M.; Terlouw, E.M.C.; Mullen, A.M.; Franco, D.; Warner, R.D.; Lorenzo, J.M.; Purslow, P.P.; Gerrard, D.; Hopkins, D.L.; Troy, D.; et al. Molecular Signatures of Beef Tenderness: Underlying Mechanisms Based on Integromics of Protein Biomarkers from Multi-Platform Proteomics Studies. Meat Sci. 2021, 172, 108311. [Google Scholar] [CrossRef] [PubMed]
- Knight, M.I.; Butler, K.L.; Linden, N.P.; Burnett, V.F.; Ball, A.J.; McDonagh, M.B.; Behrendt, R. Understanding the Impact of Sire Lean Meat Yield Breeding Value on Carcass Composition, Meat Quality, Nutrient and Mineral Content of Australian Lamb. Meat Sci. 2020, 170, 108236. [Google Scholar] [CrossRef]
- Calnan, H.B.; Jacob, R.H.; Pethick, D.W.; Gardner, G.E. Selection for Intramuscular Fat and Lean Meat Yield Will Improve the Bloomed Colour of Australian Lamb Loin Meat. Meat Sci. 2017, 131, 187–195. [Google Scholar] [CrossRef]
- Calnan, H.B.; Jacob, R.H.; Pethick, D.W.; Gardner, G.E. Factors Affecting the Colour of Lamb Meat from the Longissimus Muscle during Display: The Influence of Muscle Weight and Muscle Oxidative Capacity. Meat Sci. 2014, 96, 1049–1057. [Google Scholar] [CrossRef]
- Hopkins, D.L.; Fogarty, N.M.; Mortimer, S.I. Genetic Related Effects on Sheep Meat Quality. Small Rumin. Res. 2011, 101, 160–172. [Google Scholar] [CrossRef]
- Anderson, F.; Pannier, L.; Pethick, D.W.; Gardner, G.E. Intramuscular Fat in Lamb Muscle and the Impact of Selection for Improved Carcass Lean Meat Yield. Animal 2015, 9, 1081–1090. [Google Scholar] [CrossRef] [PubMed]
- Pannier, L.; Pethick, D.W.; Geesink, G.H.; Ball, A.J.; Jacob, R.H.; Gardner, G.E. Intramuscular Fat in the Longissimus Muscle is Reduced in Lambs from Sires Selected for Leanness. Meat Sci. 2014, 96, 1068–1075. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, S.I.; van der Werf, J.H.J.; Jacob, R.H.; Hopkins, D.L.; Pannier, L.; Pearce, K.L.; Gardner, G.E.; Warner, R.D.; Geesink, G.H.; Edwards, J.E.H.; et al. Genetic Parameters for Meat Quality Traits of Australian Lamb Meat. Meat Sci. 2014, 96, 1016–1024. [Google Scholar] [CrossRef]
- Brito, L.F.; McEwan, J.C.; Miller, S.; Bain, W.; Lee, M.; Dodds, K.; Newman, S.A.; Pickering, N.; Schenkel, F.S.; Clarke, S. Genetic Parameters for Various Growth, Carcass and Meat Quality Traits in a New Zealand Sheep Population. Small Rumin. Res. 2017, 154, 81–91. [Google Scholar] [CrossRef]
- Stratz, P.; Schiller, K.F.; Wellmann, R.; Preuss, S.; Baes, C.; Bennewitz, J. Genetic Parameter Estimates and Targeted Association Analyses of Growth, Carcass, and Meat Quality Traits in German Merinoland and Merinoland-Cross Lambs. J. Anim. Sci. 2018, 96, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Purslow, P.P.; Gagaoua, M.; Warner, R.D. Insights on Meat Quality from Combining Traditional Studies and Proteomics. Meat Sci. 2021, 174, 108423. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.H.; Yang, D.; Gong, H.S.L.; Qi, Y.X.; Sun, H.L.; Liu, Y.B.; Liu, Y.H.; Qiu, X. Multiple Omics Analysis Reveals that High Fiber Diets Promote Gluconeogenesis and Inhibit Glycolysis in Muscle. BMC Genom. 2020, 21, 660. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Lei, S.; Jiang, T.; Zeng, J.; Zhou, S.Y.; She, Y.L. Roles of lncRNAs and circRNAs in Regulating Skeletal Muscle Development. Acta Physiol. 2020, 228, e13356. [Google Scholar] [CrossRef]
- Jung, H.J.; Lee, K.P.; Kwon, K.S.; Suh, Y. MicroRNAs in Skeletal Muscle Aging: Current Issues and Perspectives. J. Gerontol. A-Biol. 2019, 74, 1008–1014. [Google Scholar] [CrossRef]
- Wang, S.S.; Jin, J.J.; Xu, Z.Y.; Zuo, B. Functions and Regulatory Mechanisms of lncRNAs in Skeletal Myogenesis, Muscle Disease and Meat Production. Cells 2019, 8, 1107. [Google Scholar] [CrossRef]
- Heller, R.C.; Chung, S.; Crissy, K.; Dumas, K.; Schuster, D.; Schoenfeld, T.W. Engineering of a Thermostable Viral Polymerase Using Metagenome-Derived Diversity for Highly Sensitive and Specific RT-PCR. Nucleic Acids Res. 2019, 47, 3619–3630. [Google Scholar] [CrossRef] [PubMed]
- Castillo, D.; Galvez, J.M.; Herrera, L.J.; San Roman, B.; Rojas, F.; Rojas, I. Integration of RNA-Seq Data With Heterogeneous Microarray Data for Breast Cancer Profiling. BMC Bioinform. 2017, 18, 506. [Google Scholar] [CrossRef] [PubMed]
- von der Haar, M.; Lindner, P.; Scheper, T.; Stahl, F. Array Analysis Manager-An Automated DNA Microarray Analysis Tool Simplifying Microarray Data Filtering, Bias Recognition, Normalization, and Expression Analysis. Eng. Life Sci. 2017, 17, 841–846. [Google Scholar] [CrossRef]
- Bolon-Canedo, V.; Alonso-Betanzos, A. Microarray Bioinformatics, 1st ed.; Humana: New York, NY, USA, 2019; pp. 283–293. [Google Scholar]
- James, K.; Cockell, S.J.; Zenkin, N. Deep Sequencing Approaches for the Analysis of Prokaryotic Transcriptional Boundaries and Dynamics. Methods 2017, 120, 76–84. [Google Scholar] [CrossRef] [PubMed]
- Jazayeri, S.M.; Munoz, L.M.M.; Romero, H.M. RNA-SEQ: A Glance at Technologies and Methodologies. Acta Biol. Colomb. 2015, 20, 23–35. [Google Scholar] [CrossRef]
- Piorkowska, K.; Malopolska, M.; Ropka-Molik, K.; Szyndler-Nedza, M.; Wiechniak, A.; Zukowski, K.; Lambert, B.; Tyra, M. Evaluation of SCD, ACACA and FASN Mutations: Effects on Pork Quality and Other Production Traits in Pigs Selected Based on RNA-Seq Results. Animals 2020, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Czerniawska-Piatkowska, E.; Kowalewska-Luczak, I.; Pecka-Kielb, E.; Banaszewska, D. Effects of FASN and SCD Gene Poly-morphism on the Composition of Sheep’s Milk. J. Anim. Plant Sci. 2021, 31, 906–912. [Google Scholar] [CrossRef]
- Jarc, E.; Petan, T. Lipid Droplets and the Management of Cellular Stress. Yale J. Biol. Med. 2019, 92, 435–452. [Google Scholar]
- Cheng, S.R.; Wang, X.Y.; Wang, Q.; Yang, L.; Shi, J.P.; Zhang, Q.W. Comparative Analysis of Longissimus Dorsi Tissue from Two Sheep Groups Identifies Differentially Expressed Genes Related to Growth, Development and Meat Quality. Genomics 2020, 112, 3322–3330. [Google Scholar] [CrossRef]
- Peng, H.Y.; Hu, M.Y.; Liu, Z.X.; Lai, W.N.; Shi, L.L.; Zhao, Z.L.; Ma, H.H.; Li, Y.M.; Yan, S.Q. Transcriptome Analysis of the Liver and Muscle Tissues of Dorper and Small-Tailed Han Sheep. Front. Genet. 2022, 13, 868717. [Google Scholar] [CrossRef]
- An, X.J.; Zhang, S.W.; Li, T.T.; Chen, N.N.; Wang, X.; Zhang, B.J.; Ma, Y.J. Transcriptomics Analysis Reveals the Effect of Broussonetia papyrifera L. Fermented Feed on Meat Quality Traits in Fattening Lamb. PeerJ 2021, 9, e11295. [Google Scholar] [CrossRef]
- Zhao, Z.D.; Raza, S.H.A.; Tian, H.S.; Shi, B.G.; Luo, Y.Z.; Wang, J.Q.; Liu, X.; Li, S.B.; Bai, Y.B.; Hu, J. Effects of Overexpression of ACSL1 Gene on the Synthesis of Unsaturated Fatty Acids in Adipocytes of Bovine. Arch. Biochem. Biophys. 2020, 695, 108648. [Google Scholar] [CrossRef]
- Xu, X.C.; Liu, T.D.; Fan, S.S.; Ma, W.P.; Chen, W.L.; Zhang, X. Effects of Fermented Caragana Korshinskii on the Intramuscular Fat Content and Expression of FABP3, UBE3C, ADRB3, LIPE, and SCD in Different Muscles of Tan Sheep. Czech J. Anim. Sci. 2020, 65, 145–152. [Google Scholar] [CrossRef]
- Zhang, M.; Guo, Y.Y.; Su, R.A.; Corazzin, M.; Hou, R.; Xie, J.Y.; Zhang, Y.; Zhao, L.H.; Su, L.; Jin, Y. Transcriptome Analysis Reveals the Molecular Regulatory Network of Muscle Development and Meat Quality in Sunit Lamb Supplemented with Dietary Probiotic. Meat Sci. 2022, 194, 108996. [Google Scholar] [CrossRef]
- Zhao, Y.B.; Zhang, Y.M.; Khas, E.; Bai, C.; Cao, Q.N.; Ao, C.J. Transcriptome Analysis Reveals Candidate Genes of the Synthesis of Branched-Chain Fatty Acids Related to Mutton Flavor in the Lamb Liver Using Allium mongolicum Regel Extract. J. Anim. Sci. 2022, 100, skac256. [Google Scholar] [CrossRef] [PubMed]
- Su, R.N.; Luo, Y.L.; Wang, B.H.; Hou, Y.R.; Zhao, L.H.; Su, L.; Yao, D.; Qian, Y.; Jin, Y. Effects of Physical Exercise on Meat Quality Characteristics of Sunit Sheep. Small Rumin. Res. 2019, 173, 54–58. [Google Scholar] [CrossRef]
- Wen, Y.L.; Li, S.B.; Bao, G.L.; Wang, J.Q.; Liu, X.; Hu, J.; Zhao, F.F.; Zhao, Z.D.; Shi, B.A.; Luo, Y.Z. Comparative Transcriptome Analysis Reveals the Mechanism Associated With Dynamic Changes in Meat Quality of the Longissimus Thoracis Muscle in Tibetan Sheep at Different Growth Stages. Front. Vet. Sci. 2022, 9, 926725. [Google Scholar] [CrossRef]
- Berry, D.P.; Conroy, S.; Hegarty, P.J.; Evans, R.D.; Pabiou, T.; Judge, M.M. Inter-Animal Genetic Variability Exist in Organoleptic Properties of Prime Beef Meat. Meat Sci. 2021, 173, 108401. [Google Scholar] [CrossRef]
- Jorrin-Novo, J.V.; Pascual, J.; Sanchez-Lucas, R.; Romero-Rodriguez, M.C.; Rodriguez-Ortega, M.J.; Lenz, C.; Valledor, L. Fourteen Years of Plant Proteomics Reflected in Proteomics: Moving from Model Species and 2DE-Based Approaches to Orphan Species and Gel-Free Platforms. Proteomics 2015, 15, 1089–1112. [Google Scholar] [CrossRef]
- Kwan, S.H.; Baie, S.; Ismail, M.N. Profiling of Proteins and Post Translational Modifications of Channa striatus Dried Meat. Curr. Proteom. 2016, 13, 9–19. [Google Scholar] [CrossRef]
- Gu, M.H.; Wei, Y.C.; Zhang, D.Q.; Liu, Y.F. iTRAQ Based Proteomic Profile Analysis for Goat Longissimus Thoracis under Repeated Freeze-Thaw Treatments. LWT-Food Sci. Technol. 2020, 134, 109934. [Google Scholar] [CrossRef]
- Jia, W.; Zhang, R.; Liu, L.; Zhu, Z.B.; Xu, M.D.; Shi, L. Molecular Mechanism of Protein Dynamic Change for Hengshan Goat Meat during Freezing Storage Based on High-Throughput Proteomics. Food Res. Int. 2021, 143, 110289. [Google Scholar] [CrossRef] [PubMed]
- Kiran, M.; Maheswarappa, N.B.; Banerjee, R.; Ch, V.; Rapole, S. Impact of Stunning before Slaughter on Expression of Skeletal Muscles Proteome in Sheep. Anim. Biotechnol. 2021, 34, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, E.; Sattari, R.; Ariaeenejad, S.; Salami, M.; Emam-Djomeh, Z.; Fotouhi, L.; Poursasan, N.; Sheibani, N.; Ghamsari, S.; Moosavi-Movahedi, A. The Impact of Slaughtering Methods on Physicochemical Characterization of Sheep Myoglobin. J. Iran. Chem. Soc. 2019, 16, 315–324. [Google Scholar] [CrossRef]
- Jia, W.; Zhang, R.; Liu, L.; Zhu, Z.B.; Mo, H.Z.; Xu, M.D.; Shi, L.; Zhang, H. Proteomics Analysis to Investigate the Impact of Diversified Thermal Processing on Meat Tenderness in Hengshan Goat meat. Meat Sci. 2022, 183, 108655. [Google Scholar] [CrossRef]
- Warner, R.D.; Wheeler, T.L.; Ha, M.; Li, X.; Bekhit, A.E.D.; Morton, J.; Vaskoska, R.; Dunshea, F.R.; Liu, R.; Purslow, P.; et al. Meat Tenderness: Advances in Biology, Biochemistry, Molecular Mechanisms and New Technologies. Meat Sci. 2022, 185, 108657. [Google Scholar] [CrossRef]
- Li, X.; Chen, L.J.; He, F.; Li, M.; Shen, Q.W.; Zhang, D.Q. A Comparative Analysis of Phosphoproteome in Ovine Muscle at Early Postmortem in Relationship to Tenderness. J. Sci. Food Agric. 2017, 97, 4571–4579. [Google Scholar] [CrossRef]
- Li, X.; Zhang, D.Q.; Ren, C.; Bai, Y.Q.; Ijaz, M.M.; Hou, C.L.; Chen, L. Effects of Protein Posttranslational Modifications on Meat Quality: A Review. Compr. Rev. Food Sci. Food Saf. 2021, 20, 289–331. [Google Scholar] [CrossRef]
- Chen, L.; Li, Z.; Li, X.; Chen, J.; Everaert, N.; Zhang, D.Q. The Effect of Sarcoplasmic Protein Phosphorylation on Glycolysis in Postmortem Ovine Muscle. Int. J. Food Sci. Technol. 2018, 53, 2714–2722. [Google Scholar] [CrossRef]
- Huang, C.Y.; Hou, C.L.; Ijaz, M.; Yan, T.J.; Li, X.; Li, Y.L.; Zhang, D.Q. Proteomics Discovery of Protein Biomarkers Linked to Meat Quality Traits in Post-Mortem Muscles: Current Trends and Future Prospects: A Review. Trends Food Sci. Technol. 2020, 105, 416–432. [Google Scholar] [CrossRef]
- Gagaoua, M.; Duffy, G.; Alvarez, C.; Burgess, C.M.; Hamill, R.; Crofton, E.; Botinestean, C.; Ferragina, A.; Cafferky, J.; Mullen, A.M.; et al. Current Research and Emerging Tools to Improve Fresh Red Meat Quality. Ir. J. Agric. Food Res. 2022, 61, 1–23. [Google Scholar] [CrossRef]
- Baharum, S.N.; Azizan, K.A. Omics Applications for Systems Biology, 1st ed.; Springer: Cham, Switzerland, 2018; pp. 51–68. [Google Scholar]
- Wang, W.T.; Sun, B.; Hu, P.; Zhou, M.; Sun, S.J.; Du, P.F.; Ru, Y.; Suvorov, A.; Li, Y.S.; Liu, Y.B.; et al. Comparison of Differential Flavor Metabolites in Meat of Lubei White Goat, Jining Gray Goat and Boer Goat. Metabolites 2019, 9, 176. [Google Scholar] [CrossRef] [PubMed]
- Kosowska, M.; Majcher, M.A.; Fortuna, T. Volatile Compounds in Meat and Meat Products. Food Sci. Technol. 2017, 37, 1–7. [Google Scholar] [CrossRef]
- You, L.Q.; Guo, Y.S.; Luo, R.M.; Fan, Y.L.; Zhang, T.G.; Hu, Q.Q.; Bo, S. Spoilage Marker Metabolites and Pathway Analysis in Chilled Tan Sheep Meat Based on GC-MS. Food Sci. Technol. Res. 2018, 24, 635–651. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Yuan, Z.; Li, F.; Yue, X. Integrating Transcriptome and Metabolome to Identify Key Genes Regulating Important Muscular Flavour Precursors in Sheep. Animal 2022, 16, 100679. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.S.; Xu, Z.Z.; Zhang, H.B.; Wang, Y.Y.; Liu, X.X.; Wang, Q.; Xue, J.L.; Zhao, Y.; Yang, S.M. Meat Differentiation between Pasture-Fed and Concentrate-Fed Sheep/Goats by Liquid Chromatography Quadrupole Time-of-Flight Mass Spectrometry Combined with Metabolomic and Lipidomic Profiling. Meat Sci. 2021, 173, 108374. [Google Scholar] [CrossRef]
- Li, J.; Yang, Y.Y.; Tang, C.H.; Yue, S.N.; Zhao, Q.Y.; Li, F.D.; Zhang, J.M. Changes in Lipids and Aroma Compounds in Intramuscular Fat from Hu Sheep. Food Chem. 2022, 383, 132611. [Google Scholar] [CrossRef]
- Li, J.; Tang, C.H.; Zhao, Q.Y.; Yang, Y.Y.; Li, F.D.; Qin, Y.C.; Liu, X.H.; Yue, X.P.; Zhang, J.M. Integrated Lipidomics and Targeted Metabolomics Analyses Reveal Changes in Flavor Precursors in Psoas Major Muscle of Castrated Lambs. Food Chem. 2020, 333, 127451. [Google Scholar] [CrossRef]
- Xu, L.; Liu, C.Y.; Li, S.B.; Xu, J.R.; Liu, H.; Zheng, X.C.; Zhang, D.Q.; Chen, L. Association of Lipidome Evolution with the Corresponding Volatile Characteristics of Postmortem Lamb during Chilled Storage. Food Res. Int. 2023, 169, 112916. [Google Scholar] [CrossRef]
- Zhang, M.; Su, R.; Corazzin, M.; Hou, R.; Zhang, Y.; Sun, L.; Hu, G.H.; Dou, L.; Guo, Y.Y.; Su, L.; et al. Lipid Transformation during Postmortem Chilled Aging in Mongolian Sheep Using Lipidomics. Food Chem. 2023, 405, 134882. [Google Scholar] [CrossRef]
- Jia, W.; Li, R.T.; Wu, X.X.; Liu, L.; Liu, S.X.; Shi, L. Molecular Mechanism of Lipid Transformation in Cold Chain Storage of Tan Sheep. Food Chem. 2021, 347, 129007. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Wu, X.X.; Li, R.T.; Liu, S.X.; Shi, L. Effect of Nisin and Potassium Sorbate Additions on Lipids and Nutritional Quality of Tan Sheep Meat. Food Chem. 2021, 365, 130535. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Shi, Q.Y.; Shi, L. Effect of Irradiation Treatment on the Lipid Composition and Nutritional Quality of Goat Meat. Food Chem. 2021, 351, 129295. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Li, R.T.; Wu, X.X.; Liu, S.X.; Shi, L. UHPLC-Q-Orbitrap HRMS-Based Quantitative Lipidomics Reveals the Chemical Changes of Phospholipids during Thermal Processing Methods of Tan Sheep Meat. Food Chem. 2021, 360, 130153. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Hui, T.; Zheng, X.C.; Li, S.B.; Wei, X.R.; Li, P.; Zhang, D.Q.; Wang, Z.Y. Characterization of Key Lipids for Binding and Generating Aroma Compounds in Roasted Mutton by UPLC-ESI-MS/MS and Orbitrap Exploris GC. Food Chem. 2022, 374, 131723. [Google Scholar] [CrossRef] [PubMed]
- Jung, G.T.; Kim, K.P.; Kim, K. How to Interpret and Integrate Multi-Omics Data at Systems Level. Anim. Cells Syst. 2020, 24, 1–7. [Google Scholar] [CrossRef]
- Buccitelli, C.; Selbach, M. mRNAs, Proteins and the Emerging Principles of Gene Expression Control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef]
- Fortelny, N.; Overall, C.M.; Pavlidis, P.; Freue, G.V.C. Can We Predict Protein from mRNA Levels? Nature 2017, 547, E19–E22. [Google Scholar] [CrossRef]
- Salovska, B.; Zhu, H.W.; Gandhi, T.; Frank, M.; Li, W.X.; Rosenberger, G.; Wu, C.D.; Germain, P.L.; Zhou, H.; Hodny, Z.; et al. Isoform-Resolved Correlation Analysis between mRNA Abundance Regulation and Protein Level Degradation. Mol. Syst. Biol. 2020, 16, e9170. [Google Scholar] [CrossRef]
- Zhao, L.M.; Li, F.D.; Zhang, X.X.; Zhang, D.Y.; Li, X.L.; Zhang, Y.K.; Zhao, Y.; Song, Q.Z.; Huang, K.; Xu, D.; et al. Integrative Analysis of Transcriptomics and Proteomics of Longissimus Thoracis of the Hu Sheep Compared with the Dorper Sheep. Meat Sci. 2022, 193, 108930. [Google Scholar] [CrossRef]
- Lim, K.S.; Lee, K.T.; Park, J.E.; Chung, W.H.; Jang, G.W.; Choi, B.H.; Hong, K.C.; Kim, T.H. Identification of Differentially Expressed Genes in Longissimus Muscle of Pigs with High and Low Intramuscular Fat Content Using RNA Sequencing. Anim. Genet. 2017, 48, 166–174. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.M.; Wang, J.; Li, H.B.; Gao, L.; Geng, J.; Ma, Z.; Liu, J.M.; Zhang, J.S.; Xie, P.G.; Chen, L. Combined Transcriptome and Proteome Analyses Reveal Differences in the Longissimus Dorsi Muscle between Kazakh Cattle and Xinjiang Brown Cattle. Anim. Biosci. 2021, 34, 1439–1450. [Google Scholar] [CrossRef]
- Windarsih, A.; Warmiko, H.D.; Indrianingsih, A.W.; Rohman, A.; Ulumuddin, Y.I.; Suratno, Y.I. Untargeted Metabolomics and Proteomics Approach Using Liquid Chromatography-Orbitrap High Resolution Mass Spectrometry to Detect Pork Adulteration in Pangasius Hypopthalmus Meat. Food Chem. 2022, 386, 132856. [Google Scholar] [CrossRef]
- Ma, D.Y.; Yu, Q.Q.; Hedrick, V.E.; Cooper, B.R.; Sobreira, T.J.P.; Oh, J.H.; Chun, H.; Kim, Y.H.B. Proteomic and Metabolomic Profiling Reveals the Involvement of Apoptosis in Meat Quality Characteristics of Ovine M. Longissimus from Different Callipyge Genotypes. Meat Sci. 2020, 166, 108140. [Google Scholar] [CrossRef]
- Zhang, B.Y.; Sun, Z.Q.; Yu, Z.; Li, H.H.; Luo, H.L.; Wang, B. Transcriptome and Targeted Metabolome Analysis Provide Insights into Bile Acids? New Roles and Mechanisms on Fat Deposition and Meat Quality in Lamb. Food Res. Int. 2022, 162, 111941. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.P.; Yang, Y.X.; Zhang, M.J.; Liu, S.; Wang, H.X.; Jiao, F.; Bao, L.J.; Lin, Z.W.; Wei, X.L.; Qian, W.; et al. Transcriptomics and Metabolomics Analysis Reveal the Anti-Oxidation and Immune Boosting Effects of Mulberry Leaves in Growing Mutton Sheep. Front. Immunol. 2023, 13, 1088850. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.W.; Yue, Y.J.; Li, J.Y.; Liu, J.B.; Yuan, C.; Guo, T.T.; Zhang, D.; Yang, B.H.; Lu, Z.K. Transcriptome-Metabolome Analysis Reveals How Sires Affect Meat Quality in Hybrid Sheep Populations. Front. Nutr. 2022, 9, 967985. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Guo, Y.S.; Luo, Y.L.; Du, M.; Yin, X.; Xu, X.C.; Zhang, G.J. Integrated Metabolomics and Transcriptome Revealed the Effect of Fermented Lycium barbarum Residue Promoting Ovis Aries Immunity. Front. Immunol. 2022, 13, 889436. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Fu, Y.; Su, T.; Wang, Y.; Soladoye, O.P.; Huang, Y.; Zhao, Z.; Zhao, Y.; Wu, W. A Role of Multi-Omics Technologies in Sheep and Goat Meats: Progress and Way Ahead. Foods 2023, 12, 4069. https://doi.org/10.3390/foods12224069
Wang J, Fu Y, Su T, Wang Y, Soladoye OP, Huang Y, Zhao Z, Zhao Y, Wu W. A Role of Multi-Omics Technologies in Sheep and Goat Meats: Progress and Way Ahead. Foods. 2023; 12(22):4069. https://doi.org/10.3390/foods12224069
Chicago/Turabian StyleWang, Jin, Yu Fu, Tianyu Su, Yupeng Wang, Olugbenga P. Soladoye, Yongfu Huang, Zhongquan Zhao, Yongju Zhao, and Wei Wu. 2023. "A Role of Multi-Omics Technologies in Sheep and Goat Meats: Progress and Way Ahead" Foods 12, no. 22: 4069. https://doi.org/10.3390/foods12224069
APA StyleWang, J., Fu, Y., Su, T., Wang, Y., Soladoye, O. P., Huang, Y., Zhao, Z., Zhao, Y., & Wu, W. (2023). A Role of Multi-Omics Technologies in Sheep and Goat Meats: Progress and Way Ahead. Foods, 12(22), 4069. https://doi.org/10.3390/foods12224069