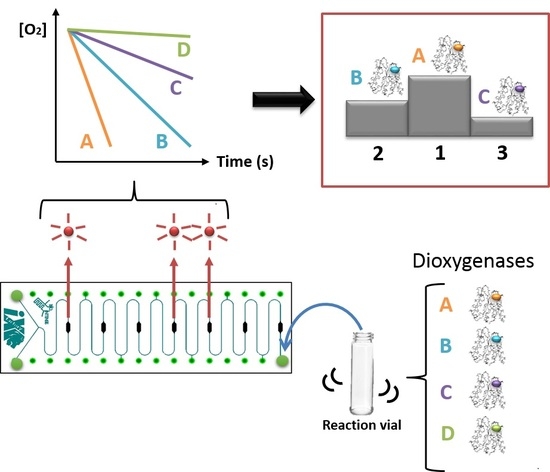
Biocatalyst Screening with a Twist: Application of Oxygen Sensors Integrated in Microchannels for Screening Whole Cell Biocatalyst Variants
Abstract
:1. Introduction
1.1. Screening Approach
1.2. Screening Technologies
Microfluidic Screening Technologies
1.3. Screening of Oxidative Biocatalysts
2. Materials and Methods
2.1. Materials
2.2. Heterologous Expression of Naphthalene Dioxygenase (in E. coli)
2.3. Preparation of Freeze-Dried Cells
2.4. Biotransformation
2.5. GC Analytical Measurement
2.6. Meander Microfluidic Channel
2.7. Oxygen Measurement Setup
3. Results
- The microreactor’s ability to distinguish the oxygen consumption due to the reaction from the cells endogenous respiration;
- The microreactor’s ability to distinguish the oxygen consumption rate of different dioxygenase variants during the reaction;
- The microreactor’s ability to differentiate the oxygen consumption rates during the reaction at various substrate concentrations.
3.1. Reaction vs. Endogenous Respiration
3.2. Variant Screening
3.3. Substrate Concentration Screening
3.4. Influence of Cell Preparation Method
4. Discussion
4.1. Measurement Setup
4.2. Reaction vs. Endogenous Respiration
4.3. Variant Screening
4.4. Substrate Concentration Screening
4.5. Influence of Cell Preparation Method
4.6. Evaluation of Oxygen Measurement in the Microchannel as a Screening Method
5. Conclusions
Supplementary Materials
Acknowledgments
Conflicts of Interest
Abbreviations
| NDO | Naphthalene dioxygenase |
| E. coli | Escherichia coli |
| GC | Gas chromatography |
| GC-FID | Gas chromatography with flame ionization detector |
| GFP | Green fluorescent protein |
| FACS | Fluorescence-activated cell sorting |
| MS | Mass spectrometry |
| HPLC | High-performance liquid chromatography |
| LC | Liquid-chromatography |
| DESI | Desorption electrospray ionization |
| NMR | Nuclear magnetic resonance |
| NIR | Near infrared |
| ROs | Rieske non-heme iron-dependent oxygenases |
| PTFE | Polytetrafluoroethylene |
| wt | Wild-type cells |
| empty | Empty-vector cells |
| pDTG141 | The NDO-containing plasmid used in this work |
References
- Adrio, J.L.; Demain, A.L. Microbial enzymes: Tools for biotechnological processes. Biomolecules 2014, 4, 117–139. [Google Scholar] [CrossRef] [PubMed]
- Burton, S.G.; Cowan, D.A.; Woodley, J.M. The search for the ideal biocatalyst. Nat. Biotechnol. 2002, 20, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Tufvesson, P.; Lima-Ramos, J.; Nordblad, M.; Woodley, J.M. Guidelines and cost analysis for catalyst production in biocatalytic processes. Org. Process Res. Dev. 2011, 15, 266–274. [Google Scholar] [CrossRef]
- Ishige, T.; Honda, K.; Shimizu, S. Whole organism biocatalysis. Curr. Opin. Chem. Biol. 2005, 9, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Ladkau, N.; Schmid, A.; Bühler, B. The microbial cell-functional unit for energy dependent multistep biocatalysis. Curr. Opin. Biotechnol. 2014, 30, 178–189. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, D.; Blank, L.M.; Schmid, A.; Bühler, B. Systems biotechnology—Rational whole-cell biocatalyst and bioprocess design. Eng. Life Sci. 2010, 10, 384–397. [Google Scholar] [CrossRef]
- Ogawa, J.; Shimizu, S. Microbial enzymes: New industrial applications from traditional screening methods. Trends Biotechnol. 1999, 17, 13–21. [Google Scholar] [CrossRef]
- Packer, M.S.; Liu, D.R. Methods for the directed evolution of proteins. Nat. Rev. Genet. 2015, 16, 379–394. [Google Scholar] [CrossRef] [PubMed]
- Doig, S.D.; Pickering, S.C.R.; Lye, G.J.; Woodley, J.M. The use of microscale processing technologies for quantification of biocatalytic Baeyer-Villiger oxidation kinetics. Biotechnol. Bioeng. 2002, 80, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Van Rossum, T.; Kengen, S.W.M.; Van Der Oost, J. Reporter-based screening and selection of enzymes. FEBS J. 2013, 280, 2979–2996. [Google Scholar] [CrossRef] [PubMed]
- Reetz, M.T.; Becker, M.H. A Method for High-Throughput Screening of Enantioselective Catalysts. Angew. Chem. Int. Ed. 1999, 38, 1758–1761. [Google Scholar] [CrossRef]
- Yan, C.; Parmeggiani, F.; Jones, E.A.; Claude, E.; Hussain, S.A.; Turner, N.J.; Flitsch, S.L.; Barran, P.E. Real-time screening of biocatalysts in live bacterial colonies. J. Am. Chem. Soc. 2017, 139, 1408–1411. [Google Scholar] [CrossRef] [PubMed]
- Heuts, D.P.H.M.; Van Hellemond, E.W.; Janssen, D.B.; Fraaije, M.W. Discovery, characterization, and kinetic analysis of an alditol oxidase from Streptomyces coelicolor. J. Biol. Chem. 2007, 282, 20283–20291. [Google Scholar] [CrossRef] [PubMed]
- Quake, S.R.; Fu, A.Y.; Spence, C.; Scherer, A.; Arnold, F.H. A microfabricated fluorescence-activated cell sorter. Nat. Biotechnol. 1999, 17, 1109–1111. [Google Scholar] [CrossRef]
- Fernández-Álvaro, E.; Snajdrova, R.; Jochens, H.; Davids, T.; Böttcher, D.; Bornscheuer, U.T. A combination of in vivo selection and cell sorting for the identification of enantioselective biocatalysts. Angew. Chem. Int. Ed. 2011, 50, 8584–8587. [Google Scholar] [CrossRef] [PubMed]
- Bornscheuer, U.T.; Huisman, G.W.; Kazlauskas, R.J.; Lutz, S.; Moore, J.C.; Robins, K. Engineering the third wave of biocatalysis. Nature 2012, 485, 185–194. [Google Scholar] [CrossRef] [PubMed]
- Parmeggiani, F.; Lovelock, S.L.; Weise, N.J.; Ahmed, S.T.; Turner, N.J. Synthesis of d- and l-Phenylalanine Derivatives by Phenylalanine Ammonia Lyases: A Multienzymatic Cascade Process. Angew. Chem. Int. Ed. 2015, 54, 4608–4611. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.; Arisawa, A.; Lin, Z.; Arnold, F.H. A high-throughput digital imaging screen for the discovery and directed evolution of oxygenases. Chem. Biol. 1999, 6, 699–706. [Google Scholar] [CrossRef]
- Schwaneberg, U.; Schmidt-Dannert, C.; Schmitt, J.; Schmid, R.D. A Continuous Spectrophotometric Assay for P450 BM-3, a Fatty Acid Hydroxylating Enzyme, and Its Mutant F87A. Anal. Biochem. 1999, 269, 359–366. [Google Scholar] [CrossRef] [PubMed]
- Duetz, W.A.; Rüedi, L.; Hermann, R.; O’Connor, K.; Büchs, J.; Witholt, B. Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 2000, 66, 2641–2646. [Google Scholar] [CrossRef] [PubMed]
- Schwaneberg, U.; Otey, C.; Cirino, P.C.; Farinas, E.; Arnold, F. Cost-Effective Whole-Cell Assay for Laboratory Evolution of Hydroxylases in Escherichia coli. J. Biomol. Screen. 2001, 6, 111–117. [Google Scholar] [CrossRef] [PubMed]
- Samorski, M.; Müller-Newen, G.; Büchs, J. Quasi-continuous combined scattered light and fluorescence measurements: A novel measurement technique for shaken microtiter plates. Biotechnol. Bioeng. 2005, 92, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Clouthier, C.M.; Pelletier, J.N. Expanding the organic toolbox: A guide to integrating biocatalysis in synthesis. Chem. Soc. Rev. 2012, 41, 1585. [Google Scholar] [CrossRef] [PubMed]
- Viefhues, M.; Sun, S.; Valikhani, D.; Nidetzky, B.; Vrouwe, E.X.; Mayr, T.; Bolivar, J.M. Tailor-made resealable micro(bio)reactors providing easy integration of in situ sensors. J. Micromech. Microeng. 2017, 27. [Google Scholar] [CrossRef]
- Couniot, N.; Francis, L.A.; Flandre, D. A 16 × 16 CMOS Capacitive Biosensor Array Towards Detection of Single Bacterial Cell. IEEE Trans. Biomed. Circuits Syst. 2016, 10, 364–374. [Google Scholar] [CrossRef] [PubMed]
- Im, H.; Shao, H.; Park, Y., Il; Peterson, V.M.; Castro, C.M.; Weissleder, R.; Lee, H. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat. Biotechnol. 2014, 490–495. [Google Scholar] [CrossRef] [PubMed]
- Wohlgemuth, R.; Plazl, I.; Žnidaršič-Plazl, P.; Gernaey, K.V.; Woodley, J.M. Microscale technology and biocatalytic processes: Opportunities and challenges for synthesis. Trends Biotechnol. 2015, 33, 302–314. [Google Scholar] [CrossRef] [PubMed]
- Abate, A.R.; Ahn, K.; Rowat, A.C.; Baret, C.; Marquez, M.; Klibanov, A.M.; Grif, A.D.; Weitz, D.A.; Aga, G.A.L. Ultrahigh-throughput screening in drop-based microfluidics for directed evolution. Proc. Natl. Acad. Sci. USA 2010, 107, 6550–6550. [Google Scholar] [CrossRef]
- Kintses, B.; Hein, C.; Mohamed, M.F.; Fischlechner, M.; Courtois, F.; Lainé, C.; Hollfelder, F. Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem. Biol. 2012, 19, 1001–1009. [Google Scholar] [CrossRef] [PubMed]
- Horka, M.; Sun, S.; Ruszczak, A.; Garstecki, P.; Mayr, T. Lifetime of Phosphorescence from Nanoparticles Yields Accurate Measurement of Concentration of Oxygen in Microdroplets, Allowing One To Monitor the Metabolism of Bacteria. Anal. Chem. 2016, 88, 12006–12012. [Google Scholar] [CrossRef] [PubMed]
- Hess, D.; Rane, A.; Demello, A.J.; Stavrakis, S. High-throughput, quantitative enzyme kinetic analysis in microdroplets using stroboscopic epifluorescence imaging. Anal. Chem. 2015, 87, 4965–4972. [Google Scholar] [CrossRef] [PubMed]
- Van Beilen, J.B.; Duetz, W.A.; Schmid, A.; Witholt, B. Practical issues in the application of oxygenases. Trends Biotechnol. 2003, 21, 170–177. [Google Scholar] [CrossRef]
- Cirino, P.C.; Arnold, F.H. Protein engineering of oxygenases for biocatalysis. Curr. Opin. Chem. Biol. 2002, 6, 130–135. [Google Scholar] [CrossRef]
- Gally, C.; Nestl, B.M.; Hauer, B. Engineering Rieske Non-Heme Iron Oxygenases for the Asymmetric Dihydroxylation of Alkenes. Angew. Chem. Int. Ed. 2015, 54, 12952–12956. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, J.W.; Orville, A.M.; Lipscomb, J.D. [14] Protocatechuate 3,4-dioxygenase from Brevibacterium fuscum. Methods Enzymol. 1990, 188, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Gibson, D.T. Stereospecific dihydroxylation of the styrene vinyl group by purified naphthalene dioxygenase from Pseudomonas sp. strain NCIB 9816-4. J. Bacteriol. 1996, 178, 3353–3356. [Google Scholar] [CrossRef] [PubMed]
- Jouanneau, Y.; Meyer, C.; Jakoncic, J.; Stojanoff, V.; Gaillard, J. Characterization of a naphthalene dioxygenase endowed with an exceptionally broad substrate specificity toward polycyclic aromatic hydrocarbons. Biochemistry 2006, 45, 12380–12391. [Google Scholar] [CrossRef] [PubMed]
- Parales, R.E.; Parales, J.V.; Gibson, D.T. Aspartate 205 in the catalytic domain of naphthalene dioxygenase is essential for activity. J. Bacteriol. 1999, 181, 1831–1837. [Google Scholar] [PubMed]
- Rachinskiy, K.; Kunze, M.; Graf, C.; Schultze, H.; Boy, M.; Büchs, J. Extension and application of the “enzyme test bench” for oxygen consuming enzyme reactions. Biotechnol. Bioeng. 2014, 111, 244–253. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Ungerböck, B.; Mayr, T. Imaging of oxygen in microreactors and microfluidic systems. Methods Appl. Fluoresc. 2015, 3, 34002. [Google Scholar] [CrossRef] [PubMed]
- Gruber, P.; Marques, M.P.C.; Szita, N.; Mayr, T. Integration and application of optical chemical sensors in microbioreactors. Lab Chip 2017. [Google Scholar] [CrossRef] [PubMed]
- Chiu, D.T.; de Mello, A.J.; Di Carlo, D.; Doyle, P.S.; Hansen, C.; Maceiczyk, R.M.; Wootton, R.C.R. Small but Perfectly Formed? Successes, Challenges, and Opportunities for Microfluidics in the Chemical and Biological Sciences. Chem 2017, 2, 201–223. [Google Scholar] [CrossRef]
- Ehgartner, J.; Sulzer, P.; Burger, T.; Kasjanow, A.; Bouwes, D.; Krühne, U.; Klimant, I.; Mayr, T. Online analysis of oxygen inside silicon-glass microreactors with integrated optical sensors. Sens. Actuators B Chem. 2016, 228, 748–757. [Google Scholar] [CrossRef] [Green Version]
- Halder, J.M. Naphthalene Dioxygenase from Pseudomonas sp. NCIB 9816-4: Systematic Analysis of the Active Site. Ph.D. Thesis, Stuttgart Universität, Stuttgart, Germany, October 2017. [Google Scholar]
- Halder, J.M.; Nestl, B.M.; Hauer, B. Semirational Engineering of the Naphthalene Dioxygenase from Pseudomonas sp. NCIB 9816-4 towards Selective Asymmetric Dihydroxylation. ChemCatChem 2018, 10, 178–182. [Google Scholar] [CrossRef]
- Parales, R.E.; Resnick, S.M.; Yu, C.L.; Boyd, D.R.; Sharma, N.D.; Gibson, D.T. Regioselectivity and enantioselectivity of naphthalene dioxygenase during arene cis-dihydroxylation: Control by Phenylalanine 352 in the alfa subunit. J. Bacteriol. 2000, 182, 5495–5504. [Google Scholar] [CrossRef] [PubMed]
- Ensley, B.D.; Ratzkin, B.J.; Osslund, T.D.; Simon, M.J.; Wackett, L.P.; Gibson, D.T. Expression of Naphthalene Oxidation Genes in Escherichia coli Results in the Biosynthesis of Indigo. Science 1983, 222, 167–169. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.; Dodge, T.C.; Pepsin, M.; Weyler, W. Application of metabolic engineering to improve both the production and use of biotech indigo. J. Ind. Microbiol. Biotechnol. 2002, 28, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Pathak, H.; Madamwar, D. Biosynthesis of indigo dye by newly isolated naphthalene-degrading strain Pseudomonas sp. HOB1 and its application in dyeing cotton fabric. Appl. Biochem. Biotechnol. 2010, 160, 1616–1626. [Google Scholar] [CrossRef] [PubMed]
- De Carvalho, C.C.C.R. Enzymatic and whole cell catalysis: Finding new strategies for old processes. Biotechnol. Adv. 2011, 29, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Wachtmeister, J.; Rother, D. Recent advances in whole cell biocatalysis techniques bridging from investigative to industrial scale. Curr. Opin. Biotechnol. 2016, 42, 169–177. [Google Scholar] [CrossRef] [PubMed]









| Biocatalyst | Substrate | Amount of Oxygenase Component of NDO | Measurement Setup | Oxygen Uptake (mM·min−1) |
|---|---|---|---|---|
| NDO from Sphingomonas CHY-1 [37] | Naphthalene (0.1 mM) | PhnI (0.13 µM) | Clark-type oxygen electrode | 14.95 |
| NDO from Pseudomonas sp. Strain NCIB 9816-4 [36] | Styrene (0.1 mM) | ISPNAP (25 µg) | 0.0700 | |
| NDO (pDTG141) wild-type in E. coli JM109/DE3) | Styrene (1 mM) | - | Luminescent oxygen sensors | 0.3202 |
| NDO variant V260A in E. coli JM109/DE3) | Styrene (2 mM) | 0.6553 | ||
| NDO variant H295A in E. coli JM109/DE3) | Styrene (2 mM) | 0.4560 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandes, A.C.; Halder, J.M.; Nestl, B.M.; Hauer, B.; Gernaey, K.V.; Krühne, U. Biocatalyst Screening with a Twist: Application of Oxygen Sensors Integrated in Microchannels for Screening Whole Cell Biocatalyst Variants. Bioengineering 2018, 5, 30. https://doi.org/10.3390/bioengineering5020030
Fernandes AC, Halder JM, Nestl BM, Hauer B, Gernaey KV, Krühne U. Biocatalyst Screening with a Twist: Application of Oxygen Sensors Integrated in Microchannels for Screening Whole Cell Biocatalyst Variants. Bioengineering. 2018; 5(2):30. https://doi.org/10.3390/bioengineering5020030
Chicago/Turabian StyleFernandes, Ana C., Julia M. Halder, Bettina M. Nestl, Bernhard Hauer, Krist V. Gernaey, and Ulrich Krühne. 2018. "Biocatalyst Screening with a Twist: Application of Oxygen Sensors Integrated in Microchannels for Screening Whole Cell Biocatalyst Variants" Bioengineering 5, no. 2: 30. https://doi.org/10.3390/bioengineering5020030
APA StyleFernandes, A. C., Halder, J. M., Nestl, B. M., Hauer, B., Gernaey, K. V., & Krühne, U. (2018). Biocatalyst Screening with a Twist: Application of Oxygen Sensors Integrated in Microchannels for Screening Whole Cell Biocatalyst Variants. Bioengineering, 5(2), 30. https://doi.org/10.3390/bioengineering5020030