Holistic Design of Experiments Using an Integrated Process Model
Abstract
:1. Introduction
1.1. Option A: Improving Model Estimates via DoE
1.2. Option B: Improving Acceptance Limits via Spiking Studies
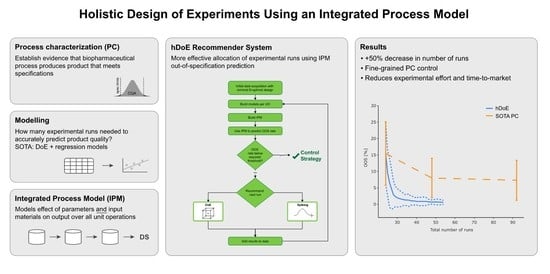
- Develop a recommender system, called the holistic design of experiments (hDoE), that suggests the optimal runs (DoE or spiking) at specific UOs that lead to the fastest increase in process understanding. Here we define process understanding as the accuracy and precision of the (unknown) true relation between all PPs and CQAs, as well as the input/output relation of individual UOs. We describe this method in Section 3.1;
- Demonstrate that using such a recommender system can lead to a significant reduction in the required number of total runs of a process characterization study (PCS) using state-of-the-art workflows. We verify this in a set of simulation studies presented in Section 3.2;
- Identify an accelerated workflow for PCS using hDoE that can be applied in practice; see Section 4.1.
2. Materials and Methods
2.1. Optimal Designs
2.2. Integrated Process Model
2.3. Simulation Study
2.3.1. Study A: Baseline
2.3.2. Study B: Load Effect Set to One
2.3.3. Study C: All Load Effects Set to One
2.3.4. Study D: Disabled Probability-Ratio-Threshold
3. Results
3.1. Holistic Design of Experiments
3.2. Simulation Results
3.2.1. Out-of-Specification Rates
3.2.2. Run Allocation
3.2.3. Parameter Estimates
4. Discussion
4.1. hDoE in Process Characterization
4.2. Outlook: Changing PP Setpoints to Increase the PAR
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- ICH. ICH Guideline Q8 (R2) on Pharmaceutical Development; EMA: London, UK, 2017. [Google Scholar]
- Burdick, R.; LeBlond, D.; Pfahler, L.; Quiroz, J.; Sidor, L.; Vukovinsky, K.; Zhang, L. Statistical Applications for Chemistry, Manufacturing and Controls (CMC) in the Pharmaceutical Industry; Springer: Cham, Switzerland, 2017. [Google Scholar]
- FDA. Process Validation: General Principles and Practices; US FDA: Rockville, MD, USA, 2011. [Google Scholar]
- Montgomery, D.C.; Peck, E.A.; Vining, G.G. Introduction to Linear Regression Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2021. [Google Scholar]
- Montgomery, D.C. Design and Analysis of Experiments; John Wiley & Sons: Hoboken, NJ, USA, 2017. [Google Scholar]
- EMA. Questions and Answers: Improving the Understanding of NORs, PARs, DSp and Normal Variability of Process Parameters; EMA: London, UK, 2017. [Google Scholar]
- Howe, W. Two-sided tolerance limits for normal populations—Some improvements. J. Am. Stat. Assoc. 1969, 64, 610–620. [Google Scholar]
- Krishnamoorthy, K.; Mathew, T. Statistical Tolerance Regions: Theory, Applications, and Computation; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Wallis, W.A. Tolerance intervals for linear regression. In Proceedings of the Second Berkeley Symposium on Mathematical Statistics and Probability, Berkeley, CA, USA, January 1951. [Google Scholar]
- Francq, B.G.; Lin, D.; Hoyer, W. Confidence, prediction, and tolerance in linear mixed models. Stat. Med. 2019, 38, 5603–5622. [Google Scholar] [CrossRef]
- Marschall, L.; Taylor, C.; Zahel, T.; Kunzelmann, M.; Wiedenmann, A.; Presser, B.; Studts, J.; Herwig, C. Specification-driven acceptance criteria for validation of biopharmaceutical processes. Front. Bioeng. Biotechnol. 2022, 10, 1010583. [Google Scholar] [CrossRef]
- Seely, R.; Munyakazi, L.; Haury, J. Statistical tools for setting in-process acceptance criteria. Dev. Biol. 2003, 113, 17–26. [Google Scholar]
- Wang, X.; Germansderfer, A.; Harms, J.; Rathore, A. Using statistical analysis for setting process validation acceptance criteria for biotech products. Biotechnol. Prog. 2007, 23, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Taylor, C.; Pretzner, B.; Zahel, T.; Herwig, C. Architectural & Technological Improvements to Integrated Bioprocess Models towards Real-Time Applications. MDPI Bioeng. 2022, 9, 534. [Google Scholar]
- Darling, A. Considerations in performing virus spiking experiments and process validation studies. Dev. Biol. Stand. 1993, 81, 221–229. [Google Scholar] [PubMed]
- Shukla, A.; Jiang, C.; Ma, J.; Rubacha, M.; Flansburg, L.; Lee, S. Demonstration of robust host cell protein clearance in biopharmaceutical downstream processes. Biotechnol. Prog. 2008, 24, 615–622. [Google Scholar] [CrossRef]
- Johnson, R.; Montgomery, D.; Jones, B. An Expository Paper on Optimal Design. Qual. Eng. 2011, 23, 287–301. [Google Scholar] [CrossRef] [Green Version]
- de Aguiar, F.; Bourguignon, B.; Khots, M.; Massart, D.; Phan-Than-Luu, R. D-optimal designs. Chemom. Intell. Lab. Syst. 1995, 30, 199–210. [Google Scholar] [CrossRef]
- Goos, P.; Jones, B.; Syafitri, U. I-optimal design of mixture experiments. J. Am. Stat. Assoc. 2016, 111, 899–911. [Google Scholar] [CrossRef]
- Jones, B.; Allen-Moyer, K.; Goos, P. A-optimal versus D-optimal design of screening experiments. J. Qual. Technol. 2021, 53, 369–382. [Google Scholar] [CrossRef]
- Velayudhan, A. Overview of integrated models for bioprocess engineering. Curr. Opin. Chem. Eng. 2014, 6, 83–89. [Google Scholar] [CrossRef]
- Mooney, C. Monte Carlo Simulation; Sage: Thousand Oaks, CA, USA, 1997. [Google Scholar]
- Hahn, G. The hazards of extrapolation in regression analysis. J. Qual. Technol. 1977, 9, 159–165. [Google Scholar] [CrossRef]
- Hamada, C.; Hamada, M. All-subsets regression under effect heredity restrictions for experimental designs with complex aliasing. Qual. Reliab. Eng. Int. 2010, 26, 75–81. [Google Scholar] [CrossRef]
- Desboulets, L.D.D. A review on variable selection in regression analysis. Econometrics 2018, 6, 45. [Google Scholar] [CrossRef] [Green Version]
- Hastings, W.K. Monte Carlo Sampling Methods Using Markov Chains and Their Applications; Oxford University Press: Oxford, UK, 1970. [Google Scholar]
- Olusegun, A.M.; Dikko, H.G.; Gulumbe, S.U. Identifying the limitation of stepwise selection for variable selection in regression analysis. Am. J. Theor. Appl. Stat. 2015, 4, 414–419. [Google Scholar] [CrossRef]
- Committee for Medicinal Products for Human Use. Process Validation for the Manufacture of Biotechnology-Derived Active Substances and Data to Be Provided in Regulatory Submissions; EMA: London, UK, 2016. [Google Scholar]
- SAS Institute Inc. JMP® 16 Profilers; SAS Institute Inc.: Cary, NC, USA, 2020–2021. [Google Scholar]
- Joyce, J. Kullback-Leibler Divergence. In International Encyclopedia of Statistical Science; Springer: Berlin/Heidelberg, Germany, 2011; pp. 720–722. [Google Scholar]










| Parameter | Value |
|---|---|
| Number of UOs | 4 |
| Parameters per UO | 5 |
| CQA type | Impurity |
| hDoE start runs | 6 |
| Noise/std ratio for residual error | 0.5 (0.9 in study D) |
| Number of runs recommended per cycle | 1 |
| Variable selection method | Bi-directional stepwise |
| p-value threshold for including effect | 0.25 |
| p-values threshold for excluding effect | 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oberleitner, T.; Zahel, T.; Pretzner, B.; Herwig, C. Holistic Design of Experiments Using an Integrated Process Model. Bioengineering 2022, 9, 643. https://doi.org/10.3390/bioengineering9110643
Oberleitner T, Zahel T, Pretzner B, Herwig C. Holistic Design of Experiments Using an Integrated Process Model. Bioengineering. 2022; 9(11):643. https://doi.org/10.3390/bioengineering9110643
Chicago/Turabian StyleOberleitner, Thomas, Thomas Zahel, Barbara Pretzner, and Christoph Herwig. 2022. "Holistic Design of Experiments Using an Integrated Process Model" Bioengineering 9, no. 11: 643. https://doi.org/10.3390/bioengineering9110643
APA StyleOberleitner, T., Zahel, T., Pretzner, B., & Herwig, C. (2022). Holistic Design of Experiments Using an Integrated Process Model. Bioengineering, 9(11), 643. https://doi.org/10.3390/bioengineering9110643