Genomic Variants and Multilevel Regulation of ABCA1, ABCG1, and SCARB1 Expression in Atherogenesis
Abstract
:1. Introduction
2. ABCA1
2.1. Expression Changes in Atherosclerosis
2.2. Studies of Overexpressing and Knockout Mice
2.3. Expression Regulation
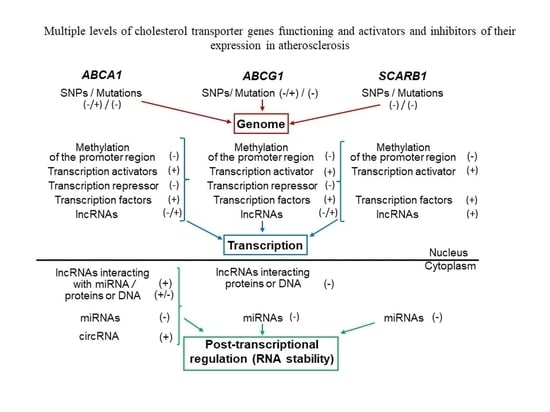
2.3.1. Changes at the Genome Level
2.3.2. Changes at the Level of Transcription Regulation
2.3.3. Changes at the Level of Post-Transcriptional Regulation of Expression
miRNAs
LncRNAs
CircRNAs
3. ABCG1
3.1. Expression Changes in Atherosclerosis
3.2. Studies of Overexpressing and Knockout Mice
3.3. Expression Regulation
3.3.1. Changes at the Genome Level
3.3.2. Changes at the Level of Transcription Regulation
3.3.3. Changes at the Level of Post-transcriptional Regulation of Expression
4. SR-BI
4.1. Expression Changes in Atherosclerosis
4.2. Studies of Overexpressing and Knockout Mice
4.3. Expression Regulation
4.3.1. Changes at the Genome Level
4.3.2. Changes at the Level of Transcription Regulation
4.3.3. Changes at the Level of Post-transcriptional Regulation of Expression
5. Medical Application of Data on the Transporter Genes Functioning in CVD
5.1. Transcriptional Regulation of Expression
5.2. Post-transcriptional Regulation of Expression
5.2.1. MiRNAs
5.2.2. LncRNAs
5.2.3. CircRNAs
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Moore, K.J.; Tabas, I. Macrophages in the pathogenesis of atherosclerosis. Cell 2011, 145, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansson, G.K.; Hermansson, A. The immune system in atherosclerosis. Nat. Immunol. 2011, 12, 204–212. [Google Scholar] [CrossRef]
- Glass, C.K.; Witztum, J.L. Atherosclerosis. the road ahead. Cell 2001, 104, 503–516. [Google Scholar] [CrossRef] [Green Version]
- Lusis, A.J. Atherosclerosis. Nature 2000, 407, 233–241. [Google Scholar] [CrossRef]
- Oram, J.F.; Heinecke, J.W. ATP-binding cassette transporter A1: A cell cholesterol exporter that protects against cardiovascular disease. Physiol. Rev. 2005, 85, 1343–1372. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, D.Y.; Savushkin, E.V.; Dergunov, A.D. Intracellular and Plasma Membrane Events in Cholesterol Transport and Homeostasis. J. Lipids 2018, 2018, 3965054. [Google Scholar] [CrossRef] [Green Version]
- Voight, B.F.; Peloso, G.M.; Orho-Melander, M.; Frikke-Schmidt, R.; Barbalic, M.; Jensen, M.K.; Hindy, G.; Holm, H.; Ding, E.L.; Johnson, T.; et al. Plasma HDL cholesterol and risk of myocardial infarction: A mendelian randomisation study. Lancet 2012, 380, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Rosenson, R.S.; Brewer, H.B., Jr.; Barter, P.J.; Bjorkegren, J.L.M.; Chapman, M.J.; Gaudet, D.; Kim, D.S.; Niesor, E.; Rye, K.A.; Sacks, F.M.; et al. HDL and atherosclerotic cardiovascular disease: Genetic insights into complex biology. Nat. Rev. Cardiol. 2018, 15, 9–19. [Google Scholar] [CrossRef]
- Rosenson, R.S.; Brewer, H.B., Jr.; Ansell, B.J.; Barter, P.; Chapman, M.J.; Heinecke, J.W.; Kontush, A.; Tall, A.R.; Webb, N.R. Dysfunctional HDL and atherosclerotic cardiovascular disease. Nat. Rev. Cardiol. 2016, 13, 48–60. [Google Scholar] [CrossRef]
- Rader, D.J.; Hovingh, G.K. HDL and cardiovascular disease. Lancet 2014, 384, 618–625. [Google Scholar] [CrossRef]
- Von Eckardstein, A.; Nofer, J.R.; Assmann, G. High density lipoproteins and arteriosclerosis. Role of cholesterol efflux and reverse cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 13–27. [Google Scholar] [CrossRef] [Green Version]
- Rosenson, R.S.; Brewer, H.B., Jr.; Davidson, W.S.; Fayad, Z.A.; Fuster, V.; Goldstein, J.; Hellerstein, M.; Jiang, X.C.; Phillips, M.C.; Rader, D.J.; et al. Cholesterol efflux and atheroprotection: Advancing the concept of reverse cholesterol transport. Circulation 2012, 125, 1905–1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, L.R.; Diniz, T.A.; Antunes, B.M.; Rossi, F.E.; Caperuto, E.C.; Lira, F.S.; Goncalves, D.C. Reverse Cholesterol Transport: Molecular Mechanisms and the Non-medical Approach to Enhance HDL Cholesterol. Front. Physiol. 2018, 9, 526. [Google Scholar] [CrossRef]
- Yvan-Charvet, L.; Wang, N.; Tall, A.R. Role of HDL, ABCA1, and ABCG1 transporters in cholesterol efflux and immune responses. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 139–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouimet, M.; Barrett, T.J.; Fisher, E.A. HDL and Reverse Cholesterol Transport. Circ. Res. 2019, 124, 1505–1518. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, D.Y.; Savushkin, E.V.; Garaeva, E.A.; Dergunov, A.D. Cholesterol Efflux and Reverse Cholesterol Transport: Experimental Approaches. Curr. Med. Chem. 2016, 23, 3883–3908. [Google Scholar] [CrossRef]
- Khera, A.V.; Cuchel, M.; Llera-Moya, M.; Rodrigues, A.; Burke, M.F.; Jafri, K.; French, B.C.; Phillips, J.A.; Mucksavage, M.L.; Wilensky, R.L.; et al. Cholesterol efflux capacity, high-density lipoprotein function, and atherosclerosis. N. Engl. J. Med. 2011, 364, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Rohatgi, A.; Khera, A.; Berry, J.D.; Givens, E.G.; Ayers, C.R.; Wedin, K.E.; Neeland, I.J.; Yuhanna, I.S.; Rader, D.R.; de Lemos, J.A.; et al. HDL cholesterol efflux capacity and incident cardiovascular events. N. Engl. J. Med. 2014, 371, 2383–2393. [Google Scholar] [CrossRef] [Green Version]
- Saleheen, D.; Scott, R.; Javad, S.; Zhao, W.; Rodrigues, A.; Picataggi, A.; Lukmanova, D.; Mucksavage, M.L.; Luben, R.; Billheimer, J.; et al. Association of HDL cholesterol efflux capacity with incident coronary heart disease events: A prospective case-control study. Lancet Diabetes Endocrinol. 2015, 3, 507–513. [Google Scholar] [CrossRef] [Green Version]
- Shea, S.; Stein, J.H.; Jorgensen, N.W.; McClelland, R.L.; Tascau, L.; Shrager, S.; Heinecke, J.W.; Yvan-Charvet, L.; Tall, A.R. Cholesterol Mass Efflux Capacity, Incident Cardiovascular Disease, and Progression of Carotid Plaque. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Fisher, E.A.; Feig, J.E.; Hewing, B.; Hazen, S.L.; Smith, J.D. High-density lipoprotein function, dysfunction, and reverse cholesterol transport. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 2813–2820. [Google Scholar] [CrossRef] [Green Version]
- Kaminski, W.E.; Piehler, A.; Wenzel, J.J. ABC A-subfamily transporters: Structure, function and disease. Biochim. Biophys. Acta 2006, 1762, 510–524. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Silver, D.L.; Costet, P.; Tall, A.R. Specific binding of ApoA-I, enhanced cholesterol efflux, and altered plasma membrane morphology in cells expressing ABC1. J. Biol. Chem. 2000, 275, 33053–33058. [Google Scholar] [CrossRef] [Green Version]
- Albrecht, C.; Soumian, S.; Amey, J.S.; Sardini, A.; Higgins, C.F.; Davies, A.H.; Gibbs, R.G. ABCA1 expression in carotid atherosclerotic plaques. Stroke 2004, 35, 2801–2806. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.F.; Cui, K.F.; Wang, J.P.; Zhang, M.; Guo, Y.P.; Li, X.Y.; Jiang, C. Significance of ABCA1 in human carotid atherosclerotic plaques. Exp. Ther. Med. 2012, 4, 297–302. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhang, J.; Zhang, S.; Yan, S.; Wang, Z.; Wang, C.; Zhang, X. MiR-30e and miR-92a are related to atherosclerosis by targeting ABCA1. Mol. Med. Rep. 2019, 19, 3298–3304. [Google Scholar] [CrossRef] [Green Version]
- Lv, Y.C.; Yin, K.; Fu, Y.C.; Zhang, D.W.; Chen, W.J.; Tang, C.K. Posttranscriptional regulation of ATP-binding cassette transporter A1 in lipid metabolism. DNA Cell Biol. 2013, 32, 348–358. [Google Scholar] [CrossRef]
- Soumian, S.; Gibbs, R.; Davies, A.; Albrecht, C. mRNA expression of genes involved in lipid efflux and matrix degradation in occlusive and ectatic atherosclerotic disease. J. Clin. Pathol. 2005, 58, 1255–1260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, S.; Arakawa, R.; Wu, C.A.; Iwamoto, N.; Lu, R.; Tsujita, M.; Abe-Dohmae, S. Calpain-mediated ABCA1 degradation: Post-translational regulation of ABCA1 for HDL biogenesis. Biochim. Biophys. Acta 2012, 1821, 547–551. [Google Scholar] [CrossRef] [PubMed]
- Demina, E.P.; Miroshnikova, V.V.; Rodygina, T.I.; Kurianov, P.S.; Vinogradov, A.G.; Denisenko, A.D.; Shvartsman, A.L. ABCA1 gene expression in peripheral blood lymphocytes and macrophages in patients with atherosclerosis. Mol. Biol. 2011, 45, 289–293. [Google Scholar] [CrossRef] [PubMed]
- Demina, E.P.; Miroshnikova, V.V.; Majorov, N.V.; Davydenko, V.V.; Shvartsman, A.L. ABCA1 mRNA and protein levels in M-CSF-activated macrophages from patients with arterial stenosis. Tsitologiia 2013, 55, 580–585. [Google Scholar] [CrossRef] [PubMed]
- Rafiei, A.; Ferns, G.A.; Ahmadi, R.; Khaledifar, A.; Rahimzadeh-Fallah, T.; Mohmmad-Rezaei, M.; Emami, S.; Bagheri, N. Expression levels of miR-27a, miR-329, ABCA1, and ABCG1 genes in peripheral blood mononuclear cells and their correlation with serum levels of oxidative stress and hs-CRP in the patients with coronary artery disease. IUBMB Life 2021, 73, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.D.; Franklin, V.; Marcel, Y.L. In vivo reverse cholesterol transport from macrophages lacking ABCA1 expression is impaired. Arterioscler. Thromb. Vasc. Biol. 2007, 27, 1837–1842. [Google Scholar] [CrossRef] [Green Version]
- Ji, A.; Meyer, J.M.; Cai, L.; Akinmusire, A.; de Beer, M.C.; Webb, N.R.; van der Westhuyzen, D.R. Scavenger receptor SR-BI in macrophage lipid metabolism. Atherosclerosis 2011, 217, 106–112. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Collins, H.L.; Ranalletta, M.; Fuki, I.V.; Billheimer, J.T.; Rothblat, G.H.; Tall, A.R.; Rader, D.J. Macrophage ABCA1 and ABCG1, but not SR-BI, promote macrophage reverse cholesterol transport in vivo. J. Clin. Investig. 2007, 117, 2216–2224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaisman, B.L.; Lambert, G.; Amar, M.; Joyce, C.; Ito, T.; Shamburek, R.D.; Cain, W.J.; Fruchart-Najib, J.; Neufeld, E.D.; Remaley, A.T.; et al. ABCA1 overexpression leads to hyperalphalipoproteinemia and increased biliary cholesterol excretion in transgenic mice. J. Clin. Investig. 2001, 108, 303–309. [Google Scholar] [CrossRef]
- Vaisman, B.L.; Demosky, S.J.; Stonik, J.A.; Ghias, M.; Knapper, C.L.; Sampson, M.L.; Dai, C.; Levine, S.J.; Remaley, A.T. Endothelial expression of human ABCA1 in mice increases plasma HDL cholesterol and reduces diet-induced atherosclerosis. J. Lipid Res. 2012, 53, 158–167. [Google Scholar] [CrossRef] [Green Version]
- Van Eck, M.; Singaraja, R.R.; Ye, D.; Hildebrand, R.B.; James, E.R.; Hayden, M.R.; Van Berkel, T.J. Macrophage ATP-binding cassette transporter A1 overexpression inhibits atherosclerotic lesion progression in low-density lipoprotein receptor knockout mice. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 929–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aiello, R.J.; Brees, D.; Bourassa, P.A.; Royer, L.; Lindsey, S.; Coskran, T.; Haghpassand, M.; Francone, O.L. Increased atherosclerosis in hyperlipidemic mice with inactivation of ABCA1 in macrophages. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 630–637. [Google Scholar] [CrossRef] [PubMed]
- Westerterp, M.; Murphy, A.J.; Wang, M.; Pagler, T.A.; Vengrenyuk, Y.; Kappus, M.S.; Gorman, D.J.; Nagareddy, P.R.; Zhu, X.; Abramowicz, S.; et al. Deficiency of ATP-binding cassette transporters A1 and G1 in macrophages increases inflammation and accelerates atherosclerosis in mice. Circ. Res. 2013, 112, 1456–1465. [Google Scholar] [CrossRef]
- Van Eck, M.; Bos, I.S.; Kaminski, W.E.; Orso, E.; Rothe, G.; Twisk, J.; Bottcher, A.; Van Amersfoort, E.S.; Christiansen-Weber, T.A.; Fung-Leung, W.P.; et al. Leukocyte ABCA1 controls susceptibility to atherosclerosis and macrophage recruitment into tissues. Proc. Natl. Acad. Sci. USA 2002, 99, 6298–6303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fredrickson, D.S.; Altrocchi, P.H.; Avioli, L.V.; Goodman, D.S.; Goodman, H.C. Tangier Disease. Combined Clinical Staff Conference at the National Institutes of Health. Ann. Intern. Med. 1961, 55, 1016–1031. [Google Scholar] [CrossRef]
- Serfaty-Lacrosniere, C.; Civeira, F.; Lanzberg, A.; Isaia, P.; Berg, J.; Janus, E.D.; Smith, M.P., Jr.; Pritchard, P.H.; Frohlich, J.; Lees, R.S. Homozygous Tangier disease and cardiovascular disease. Atherosclerosis 1994, 107, 85–98. [Google Scholar] [CrossRef]
- Lawn, R.M.; Wade, D.P.; Garvin, M.R.; Wang, X.; Schwartz, K.; Porter, J.G.; Seilhamer, J.J.; Vaughan, A.M.; Oram, J.F. The Tangier disease gene product ABC1 controls the cellular apolipoprotein-mediated lipid removal pathway. J. Clin. Investig. 1999, 104, R25–R31. [Google Scholar] [CrossRef] [Green Version]
- Rust, S.; Rosier, M.; Funke, H.; Real, J.; Amoura, Z.; Piette, J.C.; Deleuze, J.F.; Brewer, H.B.; Duverger, N.; Denefle, P.; et al. Tangier disease is caused by mutations in the gene encoding ATP-binding cassette transporter 1. Nat. Genet. 1999, 22, 352–355. [Google Scholar] [CrossRef]
- Bodzioch, M.; Orso, E.; Klucken, J.; Langmann, T.; Bottcher, A.; Diederich, W.; Drobnik, W.; Barlage, S.; Buchler, C.; Porsch-Ozcurumez, M.; et al. The gene encoding ATP-binding cassette transporter 1 is mutated in Tangier disease. Nat. Genet. 1999, 22, 347–351. [Google Scholar] [CrossRef]
- Brooks-Wilson, A.; Marcil, M.; Clee, S.M.; Zhang, L.H.; Roomp, K.; Van, D.M.; Yu, L.; Brewer, C.; Collins, J.A.; Molhuizen, H.O.; et al. Mutations in ABC1 in Tangier disease and familial high-density lipoprotein deficiency. Nat. Genet. 1999, 22, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Cameron, J.; Ranheim, T.; Halvorsen, B.; Kulseth, M.A.; Leren, T.P.; Berge, K.E. Tangier disease caused by compound heterozygosity for ABCA1 mutations R282X and Y1532C. Atherosclerosis 2010, 209, 163–166. [Google Scholar] [CrossRef]
- Maranghi, M.; Truglio, G.; Gallo, A.; Grieco, E.; Verrienti, A.; Montali, A.; Gallo, P.; Alesini, F.; Arca, M.; Lucarelli, M. A novel splicing mutation in the ABCA1 gene, causing Tangier disease and familial HDL deficiency in a large family. Biochem. Biophys. Res. Commun. 2019, 508, 487–493. [Google Scholar] [CrossRef]
- Brunham, L.R.; Kang, M.H.; Van Karnebeek, C.; Sadananda, S.N.; Collins, J.A.; Zhang, L.H.; Sayson, B.; Miao, F.; Stockler, S.; Frohlich, J.; et al. Clinical, Biochemical, and Molecular Characterization of Novel Mutations in ABCA1 in Families with Tangier Disease. JIMD Rep. 2015, 18, 51–62. [Google Scholar]
- Schaefer, E.J.; Zech, L.A.; Schwartz, D.E.; Brewer, H.B., Jr. Coronary heart disease prevalence and other clinical features in familial high-density lipoprotein deficiency (Tangier disease). Ann. Intern. Med. 1980, 93, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Clee, S.M.; Kastelein, J.J.; van Dam, M.; Marcil, M.; Roomp, K.; Zwarts, K.Y.; Collins, J.A.; Roelants, R.; Tamasawa, N.; Stulc, T.; et al. Age and residual cholesterol efflux affect HDL cholesterol levels and coronary artery disease in ABCA1 heterozygotes. J. Clin. Investig. 2000, 106, 1263–1270. [Google Scholar] [CrossRef] [Green Version]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Schnohr, P.; Steffensen, R.; Tybjaerg-Hansen, A. Mutation in ABCA1 predicted risk of ischemic heart disease in the Copenhagen City Heart Study Population. J. Am. Coll. Cardiol. 2005, 46, 1516–1520. [Google Scholar] [CrossRef] [Green Version]
- Van Dam, M.J.; de Groot, E.; Clee, S.M.; Hovingh, G.K.; Roelants, R.; Brooks-Wilson, A.; Zwinderman, A.H.; Smit, A.J.; Smelt, A.H.; Groen, A.K.; et al. Association between increased arterial-wall thickness and impairment in ABCA1-driven cholesterol efflux: An observational study. Lancet 2002, 359, 37–42. [Google Scholar] [CrossRef]
- Frikke-Schmidt, R. Genetic variation in the ABCA1 gene, HDL cholesterol, and risk of ischemic heart disease in the general population. Atherosclerosis 2010, 208, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Frikke-Schmidt, R.; Nordestgaard, B.G.; Stene, M.C.; Sethi, A.A.; Remaley, A.T.; Schnohr, P.; Grande, P.; Tybjaerg-Hansen, A. Association of loss-of-function mutations in the ABCA1 gene with high-density lipoprotein cholesterol levels and risk of ischemic heart disease. JAMA 2008, 299, 2524–2532. [Google Scholar] [CrossRef]
- Brunham, L.R.; Kastelein, J.J.; Hayden, M.R. ABCA1 gene mutations, HDL cholesterol levels, and risk of ischemic heart disease. JAMA 2008, 300, 1997–1998. [Google Scholar] [CrossRef] [PubMed]
- Panagiotakos, D.B.; Pitsavos, C.; Skoumas, J.; Chrysohoou, C.; Toutouza, M.; Stefanadis, C.I.; Toutouzas, P.K. Importance of LDL/HDL cholesterol ratio as a predictor for coronary heart disease events in patients with heterozygous familial hypercholesterolaemia: A 15-year follow-up (1987-2002). Curr. Med. Res. Opin. 2003, 19, 89–94. [Google Scholar]
- Wang, F.; Ji, Y.; Chen, X.; Song, Y.; Huang, S.; Zhou, C.; Huang, C.; Chen, Z.; Zhang, L.; Ge, J. ABCA1 variants rs2230806 (R219K), rs4149313 (M8831I), and rs9282541 (R230C) are associated with susceptibility to coronary heart disease. J. Clin. Lab. Anal. 2019, 33, e22896. [Google Scholar] [CrossRef]
- Fan, Q.; Zhu, Y.; Zhao, F. Association of rs2230806 in ABCA1 with coronary artery disease: An updated meta-analysis based on 43 research studies. Medicine 2020, 99, e18662. [Google Scholar] [CrossRef]
- Jensen, M.K.; Pai, J.K.; Mukamal, K.J.; Overvad, K.; Rimm, E.B. Common genetic variation in the ATP-binding cassette transporter A1, plasma lipids, and risk of coronary heart disease. Atherosclerosis 2007, 195, e172–e180. [Google Scholar] [CrossRef] [PubMed]
- Ghaznavi, H.; Aali, E.; Soltanpour, M.S. Association Study of the ATP—Binding Cassette Transporter A1 (ABCA1) Rs2230806 Genetic Variation with Lipid Profile and Coronary Artery Disease Risk in an Iranian Population. Open Access Maced. J. Med. Sci. 2018, 6, 274–279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, Z.; Tian, Y.; Zhao, Z.; Wu, Y.; Hu, X.; Li, J.; Chen, Q.; Wang, Y.; An, C.; Zhang, K. Association between the ABCA1 (R219K) polymorphism and lipid profiles: A meta-analysis. Sci. Rep. 2021, 11, 21718. [Google Scholar] [CrossRef]
- Zwarts, K.Y.; Clee, S.M.; Zwinderman, A.H.; Engert, J.C.; Singaraja, R.; Loubser, O.; James, E.; Roomp, K.; Hudson, T.J.; Jukema, J.W.; et al. ABCA1 regulatory variants influence coronary artery disease independent of effects on plasma lipid levels. Clin. Genet. 2002, 61, 115–125. [Google Scholar] [CrossRef]
- Lv, Y.C.; Tang, Y.Y.; Zhang, P.; Wan, W.; Yao, F.; He, P.P.; Xie, W.; Mo, Z.C.; Shi, J.F.; Wu, J.F.; et al. Histone Methyltransferase Enhancer of Zeste Homolog 2-Mediated ABCA1 Promoter DNA Methylation Contributes to the Progression of Atherosclerosis. PLoS ONE 2016, 11, e0157265. [Google Scholar]
- Ma, S.C.; Zhang, H.P.; Kong, F.Q.; Zhang, H.; Yang, C.; He, Y.Y.; Wang, Y.H.; Yang, A.N.; Tian, J.; Yang, X.L.; et al. Integration of gene expression and DNA methylation profiles provides a molecular subtype for risk assessment in atherosclerosis. Mol. Med. Rep. 2016, 13, 4791–4799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghaznavi, H.; Mahmoodi, K.; Soltanpour, M.S. A preliminary study of the association between the ABCA1 gene promoter DNA methylation and coronary artery disease risk. Mol. Biol. Res. Commun. 2018, 7, 59–65. [Google Scholar]
- Guay, S.P.; Brisson, D.; Munger, J.; Lamarche, B.; Gaudet, D.; Bouchard, L. ABCA1 gene promoter DNA methylation is associated with HDL particle profile and coronary artery disease in familial hypercholesterolemia. Epigenetics 2012, 7, 464–472. [Google Scholar] [CrossRef] [Green Version]
- Delvecchio, C.J.; Bilan, P.; Nair, P.; Capone, J.P. LXR-induced reverse cholesterol transport in human airway smooth muscle is mediated exclusively by ABCA1. Am. J. Physiol. Lung Cell. Mol. Physiol. 2008, 295, L949–L957. [Google Scholar] [CrossRef] [Green Version]
- Delvecchio, C.J.; Bilan, P.; Radford, K.; Stephen, J.; Trigatti, B.L.; Cox, G.; Parameswaran, K.; Capone, J.P. Liver X receptor stimulates cholesterol efflux and inhibits expression of proinflammatory mediators in human airway smooth muscle cells. Mol. Endocrinol. 2007, 21, 1324–1334. [Google Scholar] [CrossRef]
- Schwartz, K.; Lawn, R.M.; Wade, D.P. ABC1 gene expression and ApoA-I-mediated cholesterol efflux are regulated by LXR. Biochem. Biophys. Res. Commun. 2000, 274, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Costet, P.; Luo, Y.; Wang, N.; Tall, A.R. Sterol-dependent transactivation of the ABC1 promoter by the liver X receptor/retinoid X receptor. J. Biol. Chem. 2000, 275, 28240–28245. [Google Scholar] [CrossRef] [Green Version]
- Willy, P.J.; Umesono, K.; Ong, E.S.; Evans, R.M.; Heyman, R.A.; Mangelsdorf, D.J. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes. Dev. 1995, 9, 1033–1045. [Google Scholar] [CrossRef] [Green Version]
- Uehara, Y.; Miura, S.; Von, E.A.; Abe, S.; Fujii, A.; Matsuo, Y.; Rust, S.; Lorkowski, S.; Assmann, G.; Yamada, T.; et al. Unsaturated fatty acids suppress the expression of the ATP-binding cassette transporter G1 (ABCG1) and ABCA1 genes via an LXR/RXR responsive element. Atherosclerosis 2007, 191, 11–21. [Google Scholar] [CrossRef]
- Uehara, Y.; Engel, T.; Li, Z.; Goepfert, C.; Rust, S.; Zhou, X.; Langer, C.; Schachtrup, C.; Wiekowski, J.; Lorkowski, S.; et al. Polyunsaturated fatty acids and acetoacetate downregulate the expression of the ATP-binding cassette transporter A1. Diabetes 2002, 51, 2922–2928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Kurdi-Haidar, B.; Oram, J.F. LXR-mediated activation of macrophage stearoyl-CoA desaturase generates unsaturated fatty acids that destabilize ABCA1. J. Lipid Res. 2004, 45, 972–980. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.T.; Fu, Y.C.; Yu, W.; Lin, J.M.; Zhou, L.; Liu, L.; Wang, W. SIRT1 prevents atherosclerosis via liver-X-receptor and NF-κB signaling in a U937 cell model. Mol. Med. Rep. 2013, 8, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Nelson, J.K.; Koenis, D.S.; Scheij, S.; Cook, E.C.; Moeton, M.; Santos, A.; Lobaccaro, J.A.; Baron, S.; Zelcer, N. EEPD1 Is a Novel LXR Target Gene in Macrophages Which Regulates ABCA1 Abundance and Cholesterol Efflux. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 423–432. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daffu, G.; Shen, X.; Senatus, L.; Thiagarajan, D.; Abedini, A.; Hurtado Del, P.C.; Rosario, R.; Song, F.; Friedman, R.A.; Ramasamy, R.; et al. RAGE Suppresses ABCG1-Mediated Macrophage Cholesterol Efflux in Diabetes. Diabetes 2015, 64, 4046–4060. [Google Scholar] [CrossRef] [Green Version]
- Porsch-Ozcurumez, M.; Langmann, T.; Heimerl, S.; Borsukova, H.; Kaminski, W.E.; Drobnik, W.; Honer, C.; Schumacher, C.; Schmitz, G. The zinc finger protein 202 (ZNF202) is a transcriptional repressor of ATP binding cassette transporter A1 (ABCA1) and ABCG1 gene expression and a modulator of cellular lipid efflux. J. Biol. Chem. 2001, 276, 12427–12433. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Sallam, T.; Jones, M.; Thomas, B.J.; Wu, X.; Gilliland, T.; Qian, K.; Eskin, A.; Casero, D.; Zhang, Z.; Sandhu, J.; et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat. Med. 2018, 24, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.D.; Yao, H.H.; Wang, L.M.; Yu, M.; Shi, S.; Yuan, Z.X.; Liu, J. Knockdown of GAS5 Inhibits Atherosclerosis Progression via Reducing EZH2-Mediated ABCA1 Transcription in ApoE(-/-) Mice. Mol. Ther. Nucleic Acids 2020, 19, 84–96. [Google Scholar] [CrossRef]
- Chen, L.; Yang, W.; Guo, Y.; Chen, W.; Zheng, P.; Zeng, J.; Tong, W. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE 2017, 12, e0185406. [Google Scholar] [CrossRef]
- Peterson, S.M.; Thompson, J.A.; Ufkin, M.L.; Sathyanarayana, P.; Liaw, L.; Congdon, C.B. Common features of microRNA target prediction tools. Front. Genet. 2014, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef] [Green Version]
- Shruti, K.; Shrey, K.; Vibha, R. Micro RNAs: Tiny sequences with enormous potential. Biochem. Biophys. Res. Commun. 2011, 407, 445–449. [Google Scholar] [CrossRef]
- Finnegan, E.F.; Pasquinelli, A.E. MicroRNA biogenesis: Regulating the regulators. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 51–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jonas, S.; Izaurralde, E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015, 16, 421–433. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef]
- Ullah, S.; John, P.; Bhatti, A. MicroRNAs with a role in gene regulation and in human diseases. Mol. Biol. Rep. 2014, 41, 225–232. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villen, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Selbach, M.; Schwanhausser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Santovito, D.; Egea, V.; Bidzhekov, K.; Natarelli, L.; Mourao, A.; Blanchet, X.; Wichapong, K.; Aslani, M.; Brunsen, C.; Horckmans, M.; et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci. Transl. Med. 2020, 12, eaaz2294. [Google Scholar] [CrossRef]
- Yang, D.; Wan, X.; Dennis, A.T.; Bektik, E.; Wang, Z.; Costa, M.G.S.; Fagnen, C.; Venien-Bryan, C.; Xu, X.; Gratz, D.H.; et al. MicroRNA Biophysically Modulates Cardiac Action Potential by Direct Binding to Ion Channel. Circulation 2021, 143, 1597–1613. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; MacRae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Liang, B.; Wang, X.; Song, X.; Bai, R.; Yang, H.; Yang, Z.; Xiao, C.; Bian, Y. MicroRNA-20a/b regulates cholesterol efflux through post-transcriptional repression of ATP-binding cassette transporter A1. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1862, 929–938. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Sun, Y.N.; Zhou, Y.T.; Zhang, C.L.; Lu, F.; Liu, J.; Shang, X.M. Screening and identification of microRNA involved in unstable angina using gene-chip analysis. Exp. Ther. Med. 2016, 12, 2716–2722. [Google Scholar] [CrossRef] [Green Version]
- DAmore, S.; Hardfeldt, J.; Cariello, M.; Graziano, G.; Copetti, M.; Di Tullio, G.; Piglionica, M.; Scialpi, N.; Sabba, C.; Palasciano, G.; et al. Identification of miR-9-5p as direct regulator of ABCA1 and HDL-driven reverse cholesterol transport in circulating CD14+ cells of patients with metabolic syndrome. Cardiovasc. Res. 2018, 114, 1154–1164. [Google Scholar] [CrossRef]
- Bidzhekov, K.; Gan, L.; Denecke, B.; Rostalsky, A.; Hristov, M.; Koeppel, T.A.; Zernecke, A.; Weber, C. microRNA expression signatures and parallels between monocyte subsets and atherosclerotic plaque in humans. Thromb. Haemost. 2012, 107, 619–625. [Google Scholar]
- Wang, D.; Xia, M.; Yan, X.; Li, D.; Wang, L.; Xu, Y.; Jin, T.; Ling, W. Gut microbiota metabolism of anthocyanin promotes reverse cholesterol transport in mice via repressing miRNA-10b. Circ. Res. 2012, 111, 967–981. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Wang, W.; Lin, W.; Yang, W.; Zhang, P.; Chen, M.; Ding, D.; Liu, C.; Zheng, J.; Ling, W. Apoptotic cell induction of miR-10b in macrophages contributes to advanced atherosclerosis progression in ApoE-/- mice. Cardiovasc. Res. 2018, 114, 1794–1805. [Google Scholar] [CrossRef]
- Tan, L.; Liu, L.; Jiang, Z.; Hao, X. Inhibition of microRNA-17-5p reduces the inflammation and lipid accumulation, and up-regulates ATP-binding cassette transporterA1 in atherosclerosis. J. Pharmacol. Sci. 2019, 139, 280–288. [Google Scholar] [CrossRef]
- Ren, J.; Zhang, J.; Xu, N.; Han, G.; Geng, Q.; Song, J.; Li, S.; Zhao, J.; Chen, H. Signature of circulating microRNAs as potential biomarkers in vulnerable coronary artery disease. PLoS ONE 2013, 8, e80738. [Google Scholar] [CrossRef] [Green Version]
- Xue, S.; Liu, D.; Zhu, W.; Su, Z.; Zhang, L.; Zhou, C.; Li, P. Circulating MiR-17-5p, MiR-126-5p and MiR-145-3p Are Novel Biomarkers for Diagnosis of Acute Myocardial Infarction. Front. Physiol. 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patterson, A.J.; Song, M.A.; Choe, D.; Xiao, D.; Foster, G.; Zhang, L. Early Detection of Coronary Artery Disease by Micro-RNA Analysis in Asymptomatic Patients Stratified by Coronary CT Angiography. Diagnostics 2020, 10, 875. [Google Scholar] [CrossRef]
- Liu, F.; Li, R.; Zhang, Y.; Qiu, J.; Ling, W. Association of plasma MiR-17-92 with dyslipidemia in patients with coronary artery disease. Medicine 2014, 93, e98. [Google Scholar] [CrossRef]
- Wakabayashi, I.; Eguchi, R.; Sotoda, Y.; von Lewinski, D.; Sourij, H.; Daimon, T.; Groschner, K.; Rainer, P.P. Blood levels of microRNAs associated with ischemic heart disease differ between Austrians and Japanese: A pilot study. Sci. Rep. 2020, 10, 13628. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Xu, L.; Hu, Q.; Yang, S.; Zhang, B.; Jiang, H. MiR-17-5p as circulating biomarkers for the severity of coronary atherosclerosis in coronary artery disease. Int. J. Cardiol. 2015, 197, 123–124. [Google Scholar] [CrossRef] [PubMed]
- Fichtlscherer, S.; De Rosa, S.; Fox, H.; Schwietz, T.; Fischer, A.; Liebetrau, C.; Weber, M.; Hamm, C.W.; Roxe, T.; Muller-Ardogan, M.; et al. Circulating microRNAs in patients with coronary artery disease. Circ. Res. 2010, 107, 677–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Hao, J.; Sun, X.; Zhang, Y.; Wei, Q. Circulating pro-angiogenic micro-ribonucleic acid in patients with coronary heart disease. Interact. Cardiovasc. Thorac. Surg. 2018, 27, 336–342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Huang, Y.; Lu, X.; Lu, M.; Huang, X.; Li, W.; Jiang, M. Identification and characteristics of microRNAs with altered expression patterns in a rat model of abdominal aortic aneurysms. Tohoku J. Exp. Med. 2010, 222, 187–193. [Google Scholar] [CrossRef]
- Parahuleva, M.S.; Lipps, C.; Parviz, B.; Holschermann, H.; Schieffer, B.; Schulz, R.; Euler, G. MicroRNA expression profile of human advanced coronary atherosclerotic plaques. Sci. Rep. 2018, 8, 7823. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, S.; Wu, M.; Chi, C.; Hu, D.; Cui, Y.; Song, J.; Lee, C.; Chen, H. Early diagnostic value of circulating microRNAs in patients with suspected acute myocardial infarction. J. Cell. Physiol. 2019, 234, 13649–13658. [Google Scholar] [CrossRef]
- Li, S.; Ren, J.; Xu, N.; Zhang, J.; Geng, Q.; Cao, C.; Lee, C.; Song, J.; Li, J.; Chen, H. MicroRNA-19b functions as potential anti-thrombotic protector in patients with unstable angina by targeting tissue factor. J. Mol. Cell. Cardiol. 2014, 75, 49–57. [Google Scholar] [CrossRef]
- Lv, Y.C.; Tang, Y.Y.; Peng, J.; Zhao, G.J.; Yang, J.; Yao, F.; Ouyang, X.P.; He, P.P.; Xie, W.; Tan, Y.L.; et al. MicroRNA-19b promotes macrophage cholesterol accumulation and aortic atherosclerosis by targeting ATP-binding cassette transporter A1. Atherosclerosis 2014, 236, 215–226. [Google Scholar] [CrossRef]
- Lv, Y.C.; Yang, J.; Yao, F.; Xie, W.; Tang, Y.Y.; Ouyang, X.P.; He, P.P.; Tan, Y.L.; Li, L.; Zhang, M.; et al. Diosgenin inhibits atherosclerosis via suppressing the MiR-19b-induced downregulation of ATP-binding cassette transporter A1. Atherosclerosis 2015, 240, 80–89. [Google Scholar] [CrossRef]
- Kin, K.; Miyagawa, S.; Fukushima, S.; Shirakawa, Y.; Torikai, K.; Shimamura, K.; Daimon, T.; Kawahara, Y.; Kuratani, T.; Sawa, Y. Tissue- and plasma-specific MicroRNA signatures for atherosclerotic abdominal aortic aneurysm. J. Am. Heart Assoc. 2012, 1, e000745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meder, B.; Keller, A.; Vogel, B.; Haas, J.; Sedaghat-Hamedani, F.; Kayvanpour, E.; Just, S.; Borries, A.; Rudloff, J.; Leidinger, P.; et al. MicroRNA signatures in total peripheral blood as novel biomarkers for acute myocardial. Infarct. Basic Res. Cardiol. 2011, 106, 13–23. [Google Scholar] [CrossRef]
- Stather, P.W.; Sylvius, N.; Wild, J.B.; Choke, E.; Sayers, R.D.; Bown, M.J. Differential microRNA expression profiles in peripheral arterial disease. Circ. Cardiovasc. Genet. 2013, 6, 490–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Ye, Z.M.; Chen, S.; Luo, X.Y.; Chen, S.L.; Mao, L.; Li, Y.; Jin, H.; Yu, C.; Xiang, F.X.; et al. MicroRNA-23a-5p promotes atherosclerotic plaque progression and vulnerability by repressing ATP-binding cassette transporter A1/G1 in macrophages. J. Mol. Cell. Cardiol. 2018, 123, 139–149. [Google Scholar] [CrossRef] [PubMed]
- Babaee, M.; Chamani, E.; Ahmadi, R.; Bahreini, E.; Balouchnejadmojarad, T.; Nahrkhalaji, A.S.; Fallah, S. The expression levels of miRNAs- 27a and 23a in the peripheral blood mononuclear cells (PBMCs) and their correlation with FOXO1 and some inflammatory and anti-inflammatory cytokines in the patients with coronary artery disease (CAD). Life Sci. 2020, 256, 117898. [Google Scholar] [CrossRef]
- Satoh, M.; Nasu, T.; Takahashi, Y.; Osaki, T.; Hitomi, S.; Morino, Y.; Nakamura, M. Expression of miR-23a induces telomere shortening and is associated with poor clinical outcomes in patients with coronary artery disease. Clin. Sci. 2017, 131, 2007–2017. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; He, W.; Wang, C. MiR-23a Regulates the Vasculogenesis of Coronary Artery Disease by Targeting Epidermal Growth Factor Receptor. Cardiovasc. Ther. 2016, 34, 199–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, H.; Qu, G.; Han, C.; Wang, Y.; Sun, T.; Li, F.; Wang, J.; Luo, S. MiR-34a, miR-21 and miR-23a as potential biomarkers for coronary artery disease: A pilot microarray study and confirmation in a 32 patient cohort. Exp. Mol. Med. 2015, 47, e138. [Google Scholar] [CrossRef] [Green Version]
- Jia, L.; Hao, F.; Wang, W.; Qu, Y. Circulating miR-145 is associated with plasma high-sensitivity C-reactive protein in acute ischemic stroke patients. Cell Biochem. Funct. 2015, 33, 314–319. [Google Scholar] [CrossRef]
- Wang, M.; Li, L.; Liu, R.; Song, Y.; Zhang, X.; Niu, W.; Kumar, A.K.; Guo, Z.; Hu, Z. Obesity-induced overexpression of miRNA-24 regulates cholesterol uptake and lipid metabolism by targeting SR-B1. Gene 2018, 668, 196–203. [Google Scholar] [CrossRef]
- Gecys, D.; Tatarunas, V.; Veikutiene, A.; Lesauskaite, V. New potential modulators of CYP4F2 enzyme activity in angina pectoris: Hsa-miR-24-3p and hsa-miR-34a-5p. Biomarkers 2020, 25, 40–47. [Google Scholar] [CrossRef]
- Dong, J.; Liang, Y.Z.; Zhang, J.; Wu, L.J.; Wang, S.; Hua, Q.; Yan, Y.X. Potential Role of Lipometabolism-Related MicroRNAs in Peripheral Blood Mononuclear Cells as Biomarkers for Coronary Artery Disease. J. Atheroscler. Thromb. 2017, 24, 430–441. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Li, Y.; Liu, G.; Qi, X.; Cao, X. MicroRNA-24 inhibits the proliferation and migration of endothelial cells in patients with atherosclerosis by targeting importin-O±3 and regulating inflammatory responses. Exp. Ther. Med. 2018, 15, 338–344. [Google Scholar] [PubMed] [Green Version]
- de Gonzalo-Calvo, D.; Cenarro, A.; Garlaschelli, K.; Pellegatta, F.; Vilades, D.; Nasarre, L.; Camino-Lopez, S.; Crespo, J.; Carreras, F.; Leta, R.; et al. Translating the microRNA signature of microvesicles derived from human coronary artery smooth muscle cells in patients with familial hypercholesterolemia and coronary artery disease. J. Mol. Cell. Cardiol. 2017, 106, 55–67. [Google Scholar] [CrossRef]
- Ren, K.; Zhu, X.; Zheng, Z.; Mo, Z.C.; Peng, X.S.; Zeng, Y.Z.; Ou, H.X.; Zhang, Q.H.; Qi, H.Z.; Zhao, G.J.; et al. MicroRNA-24 aggravates atherosclerosis by inhibiting selective lipid uptake from HDL cholesterol via the post-transcriptional repression of scavenger receptor class B type I. Atherosclerosis 2018, 270, 57–67. [Google Scholar] [CrossRef]
- Xue, S.; Zhu, W.; Liu, D.; Su, Z.; Zhang, L.; Chang, Q.; Li, P. Circulating miR-26a-1, miR-146a and miR-199a-1 are potential candidate biomarkers for acute myocardial infarction. Mol. Med. 2019, 25, 18. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, J.; Xie, J.; Wei, W.; Chen, M.; Zhao, X. MiR-26 controls LXR-dependent cholesterol efflux by targeting ABCA1 and ARL7. FEBS Lett. 2012, 586, 1472–1479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Cao, H.; Zhuang, J.; Wan, J.; Guan, M.; Yu, B.; Li, X.; Zhang, W. Identification of miR-130a, miR-27b and miR-210 as serum biomarkers for atherosclerosis obliterans. Clin. Chim. Acta Int. J. Clin. Chem. 2011, 412, 66–70. [Google Scholar] [CrossRef] [PubMed]
- Plana, E.; Galvez, L.; Medina, P.; Navarro, S.; Fornes-Ferrer, V.; Panadero, J.; Miralles, M. Identification of Novel microRNA Profiles Dysregulated in Plasma and Tissue of Abdominal Aortic Aneurysm Patients. Int. J. Mol. Sci. 2020, 21, 4600. [Google Scholar] [CrossRef]
- Vickers, K.C.; Shoucri, B.M.; Levin, M.G.; Wu, H.; Pearson, D.S.; Osei-Hwedieh, D.; Collins, F.S.; Remaley, A.T.; Sethupathy, P. MicroRNA-27b is a regulatory hub in lipid metabolism and is altered in dyslipidemia. Hepatology 2013, 57, 533–542. [Google Scholar] [CrossRef] [Green Version]
- Vegter, E.L.; Ovchinnikova, E.S.; van Veldhuisen, D.J.; Jaarsma, T.; Berezikov, E.; van der Meer, P.; Voors, A.A. Low circulating microRNA levels in heart failure patients are associated with atherosclerotic disease and cardiovascular-related rehospitalizations. Clin. Res. Cardiol. 2017, 106, 598–609. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Wu, J.F.; Chen, W.J.; Tang, S.L.; Mo, Z.C.; Tang, Y.Y.; Li, Y.; Wang, J.L.; Liu, X.Y.; Peng, J.; et al. MicroRNA-27a/b regulates cellular cholesterol efflux, influx and esterification/hydrolysis in THP-1 macrophages. Atherosclerosis 2014, 234, 54–64. [Google Scholar] [CrossRef]
- Goedeke, L.; Rotllan, N.; Ramirez, C.M.; Aranda, J.F.; Canfran-Duque, A.; Araldi, E.; Fernandez-Hernando, A.; Langhi, C.; de Cabo, R.; Baldan, A.; et al. miR-27b inhibits LDLR and ABCA1 expression but does not influence plasma and hepatic lipid levels in mice. Atherosclerosis 2015, 243, 499–509. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Liu, X.Q.; Liu, Y.; Sun, Y.N.; Li, S.; Li, C.M.; Li, J.; Tian, W.; Shang, X.M.; Zhou, Y.T. MicroRNA 28-5p regulates ATP-binding cassette transporter A1 via inhibiting extracellular signal-regulated kinase 2. Mol. Med. Rep. 2016, 13, 433–440. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liu, Y.; Sun, Y.N.; Li, S.; Liu, X.Q.; Li, J.; Li, C.M.; Tian, W.; Zhou, Y.T.; Shang, X.M. miR-28-5p Involved in LXR-ABCA1 Pathway is Increased in the Plasma of Unstable Angina Patients. Heart Lung Circ. 2015, 24, 724–730. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Lin, Y.; Yan, W.; Sun, Z.; Jiang, Z.; Shen, B.; Jiang, X.; Shi, J. Novel Biomarker MicroRNAs for Subtyping of Acute Coronary Syndrome: A Bioinformatics Approach. BioMed Res. Int. 2016, 2016, 4618323. [Google Scholar] [CrossRef] [Green Version]
- Bogucka-Kocka, A.; Zalewski, D.P.; Ruszel, K.P.; Stepniewski, A.; Galkowski, D.; Bogucki, J.; Komsta, L.; Kolodziej, P.; Zubilewicz, T.; Feldo, M.; et al. Dysregulation of MicroRNA Regulatory Network in Lower Extremities Arterial Disease. Front. Genet. 2019, 10, 1200. [Google Scholar] [CrossRef] [Green Version]
- Weber, M.; Baker, M.B.; Patel, R.S.; Quyyumi, A.A.; Bao, G.; Searles, C.D. MicroRNA Expression Profile in CAD Patients and the Impact of ACEI/ARB. Cardiol. Res. Pract. 2011, 2011, 532915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Xu, Y.; Zhu, Y.; Sun, H.; Juguilon, C.; Li, F.; Fan, D.; Yin, L.; Zhang, Y. Macrophage miR-34a Is a Key Regulator of Cholesterol Efflux and Atherosclerosis. Mol. Am. Soc. Gene Ther. 2020, 28, 202–216. [Google Scholar] [CrossRef]
- Raitoharju, E.; Lyytikainen, L.P.; Levula, M.; Oksala, N.; Mennander, A.; Tarkka, M.; Klopp, N.; Illig, T.; Kahonen, M.; Karhunen, P.J.; et al. miR-21, miR-210, miR-34a, and miR-146a/b are up-regulated in human atherosclerotic plaques in the Tampere Vascular Study. Atherosclerosis 2011, 219, 211–217. [Google Scholar] [CrossRef]
- Zhong, Z.; Zhong, W.; Zhang, Q.; Zhang, Q.; Yu, Z.; Wu, H. Circulating microRNA expression profiling and bioinformatics analysis of patients with coronary artery disease by RNA sequencing. J. Clin. Lab. Anal. 2020, 34, e23020. [Google Scholar] [CrossRef]
- Widmer, R.J.; Chung, W.Y.; Herrmann, J.; Jordan, K.L.; Lerman, L.O.; Lerman, A. The association between circulating microRNA levels and coronary endothelial function. PLoS ONE 2014, 9, e109650. [Google Scholar] [CrossRef] [Green Version]
- Niculescu, L.S.; Simionescu, N.; Sanda, G.M.; Carnuta, M.G.; Stancu, C.S.; Popescu, A.C.; Popescu, M.R.; Vlad, A.; Dimulescu, D.R.; Simionescu, M.; et al. MiR-486 and miR-92a Identified in Circulating HDL Discriminate between Stable and Vulnerable Coronary Artery Disease Patients. PLoS ONE 2015, 10, e0140958. [Google Scholar] [CrossRef]
- Huang, Y.; Tang, S.; Ji-Yan, C.; Huang, C.; Li, J.; Cai, A.P.; Feng, Y.Q. Circulating miR-92a expression level in patients with essential hypertension: A potential marker of atherosclerosis. J. Hum. Hypertens. 2017, 31, 200–205. [Google Scholar] [CrossRef]
- Chen, G.; Gao, J.; Sheng, Y.; Han, X.; Ji, X.; Zhao, M.; Wu, J. Diagnostic value of miR-92a in asymptomatic carotid artery stenosis patients and its ability to predict cerebrovascular events. Diagn. Pathol. 2020, 15, 74. [Google Scholar] [CrossRef]
- Faccini, J.; Ruidavets, J.B.; Cordelier, P.; Martins, F.; Maoret, J.J.; Bongard, V.; Ferrieres, J.; Roncalli, J.; Elbaz, M.; Vindis, C. Circulating miR-155, miR-145 and let-7c as diagnostic biomarkers of the coronary artery disease. Sci. Rep. 2017, 7, 42916. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Wang, H.Y.; Li, Y.; Guo, S.H.; Zhang, L.; Cai, J.H. Peripheral blood miRNAs as a biomarker for chronic cardiovascular diseases. Sci. Rep. 2014, 4, 5026. [Google Scholar] [CrossRef] [Green Version]
- Barbalata, T.; Moraru, O.E.; Stancu, C.S.; Devaux, Y.; Simionescu, M.; Sima, A.V.; Niculescu, L.S. Increased miR-142 Levels in Plasma and Atherosclerotic Plaques from Peripheral Artery Disease Patients with Post-Surgery Cardiovascular Events. Int. J. Mol. Sci. 2020, 21, 9600. [Google Scholar] [CrossRef] [PubMed]
- Loyer, X.; Potteaux, S.; Vion, A.C.; Guerin, C.L.; Boulkroun, S.; Rautou, P.E.; Ramkhelawon, B.; Esposito, B.; Dalloz, M.; Paul, J.L.; et al. Inhibition of microRNA-92a prevents endothelial dysfunction and atherosclerosis in mice. Circ. Res. 2014, 114, 434–443. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Infante, T.; Forte, E.; Punzo, B.; Cademartiri, F.; Cavaliere, C.; Soricelli, A.; Salvatore, M.; Napoli, C. Correlation of Circulating miR-765, miR-93-5p, and miR-433-3p to Obstructive Coronary Heart Disease Evaluated by Cardiac Computed Tomography. Am. J. Cardiol. 2019, 124, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, J.F.O.; Neylon, A.; McGorrian, C.; Blake, G.J. miRNA-93-5p and other miRNAs as predictors of coronary artery disease and STEMI. Int. J. Cardiol. 2016, 224, 310–316. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Lin, L.; Cao, J.; Mao, X.; Qu, Y.; Xi, B. Up-regulated miR-93 contributes to coronary atherosclerosis pathogenesis through targeting ABCA1. Int. J. Clin. Exp. Med. 2015, 8, 674–681. [Google Scholar]
- Wang, L.; Jia, X.J.; Jiang, H.J.; Du, Y.; Yang, F.; Si, S.Y.; Hong, B. MicroRNAs 185, 96, and 223 repress selective high-density lipoprotein cholesterol uptake through posttranscriptional inhibition. Mol. Cell. Biol. 2013, 33, 1956–1964. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Lei, J.; Lei, H.; Ruan, X.; Liu, Q.; Chen, Y.; Huang, W. MicroRNA-101 overexpression by IL-6 and TNF-α inhibits cholesterol efflux by suppressing ATP-binding cassette transporter A1 expression. Exp. Cell Res. 2015, 336, 33–42. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yoon, H.; Ramirez, C.M.; Lee, S.M.; Hoe, H.S.; Fernandez-Hernando, C.; Kim, J. MiR-106b impairs cholesterol efflux and increases Aβ levels by repressing ABCA1 expression. Exp. Neurol. 2012, 235, 476–483. [Google Scholar] [CrossRef] [Green Version]
- Hao, L.; Wang, X.G.; Cheng, J.D.; You, S.Z.; Ma, S.H.; Zhong, X.; Quan, L.; Luo, B. The up-regulation of endothelin-1 and down-regulation of miRNA-125a-5p, -155, and -199a/b-3p in human atherosclerotic coronary artery. Cardiovasc. Pathol. 2014, 23, 217–223. [Google Scholar] [CrossRef]
- Zhang, X.; Shao, S.; Geng, H.; Yu, Y.; Wang, C.; Liu, Z.; Yu, C.; Jiang, X.; Deng, Y.; Gao, L.; et al. Expression profiles of six circulating microRNAs critical to atherosclerosis in patients with subclinical hypothyroidism: A clinical study. J. Clin. Endocrinol. Metab. 2014, 99, E766–E774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Z.; Shen, W.J.; Kraemer, F.B.; Azhar, S. MicroRNAs 125a and 455 repress lipoprotein-supported steroidogenesis by targeting scavenger receptor class B type I in steroidogenic cells. Mol. Cell. Biol. 2012, 32, 5035–5045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adlakha, Y.K.; Khanna, S.; Singh, R.; Singh, V.P.; Agrawal, A.; Saini, N. Pro-apoptotic miRNA-128-2 modulates ABCA1, ABCG1 and RXRα expression and cholesterol homeostasis. Cell Death Dis. 2013, 4, e780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chandra, A.; Sharma, K.; Pratap, K.; Singh, V.; Saini, N. Inhibition of microRNA-128-3p attenuates hypercholesterolemia in mouse model. Life Sci. 2021, 264, 118633. [Google Scholar] [CrossRef] [PubMed]
- Wagschal, A.; Najafi-Shoushtari, S.H.; Wang, L.; Goedeke, L.; Sinha, S.; de Lemos, A.S.; Black, J.C.; Ramirez, C.M.; Li, Y.; Tewhey, R.; et al. Genome-wide identification of microRNAs regulating cholesterol and triglyceride homeostasis. Nat. Med. 2015, 21, 1290–1297. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Zhang, Z.; Zhang, L.; Chen, S.; Guo, Y.; Hong, Y. miR-143 and miR-145 promote hypoxia-induced proliferation and migration of pulmonary arterial smooth muscle cells through regulating ABCA1 expression. Cardiovasc. Pathol. 2018, 37, 15–25. [Google Scholar] [CrossRef]
- Sala, F.; Aranda, J.F.; Rotllan, N.; Ramirez, C.M.; Aryal, B.; Elia, L.; Condorelli, G.; Catapano, A.L.; Fernandez-Hernando, C.; Norata, G.D. MiR-143/145 deficiency attenuates the progression of atherosclerosis in Ldlr-/-mice. Thromb. Haemost. 2014, 112, 796–802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorur, A.; Celik, A.; Yildirim, D.D.; Gundes, A.; Tamer, L. Investigation of possible effects of microRNAs involved in regulation of lipid metabolism in the pathogenesis of atherosclerosis. Mol. Biol. Rep. 2019, 46, 909–920. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Luo, L.; Wei, X.; Gong, D.; Jin, L. Altered Plasma miR-144 as a Novel Biomarker for Coronary Artery Disease. Ann. Clin. Lab. Sci. 2018, 48, 440–445. [Google Scholar] [PubMed]
- Huesca-Gomez, C.; Torres-Paz, Y.E.; Martinez-Alvarado, R.; Fuentevilla-Alvarez, G.; Del Valle-Mondragon, L.; Torres-Tamayo, M.; Soto, M.E.; Gamboa, R. Association between the transporters ABCA1/G1 and the expression of miR-33a/144 and the carotid intima media thickness in patients with arterial hypertension. Mol. Biol. Rep. 2020, 47, 1321–1329. [Google Scholar] [CrossRef]
- Hu, Y.W.; Hu, Y.R.; Zhao, J.Y.; Li, S.F.; Ma, X.; Wu, S.G.; Lu, J.B.; Qiu, Y.R.; Sha, Y.H.; Wang, Y.C.; et al. An agomir of miR-144-3p accelerates plaque formation through impairing reverse cholesterol transport and promoting pro-inflammatory cytokine production. PLoS ONE 2014, 9, e94997. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, C.M.; Rotllan, N.; Vlassov, A.V.; Davalos, A.; Li, M.; Goedeke, L.; Aranda, J.F.; Cirera-Salinas, D.; Araldi, E.; Salerno, A.; et al. Control of cholesterol metabolism and plasma high-density lipoprotein levels by microRNA-144. Circ. Res. 2013, 112, 1592–1601. [Google Scholar] [CrossRef]
- de Aguiar Vallim, T.Q.; Tarling, E.J.; Kim, T.; Civelek, M.; Baldan, A.; Esau, C.; Edwards, P.A. MicroRNA-144 regulates hepatic ATP binding cassette transporter A1 and plasma high-density lipoprotein after activation of the nuclear receptor farnesoid X receptor. Circ. Res. 2013, 112, 1602–1612. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.H.; Zhang, L.H.; Wijesekara, N.; de Haan, W.; Butland, S.; Bhattacharjee, A.; Hayden, M.R. Regulation of ABCA1 protein expression and function in hepatic and pancreatic islet cells by miR-145. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2724–2732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Q.; Wang, F.; Honda, T.; Greis, K.D.; Redington, A.N. Ablation of miR-144 increases vimentin expression and atherosclerotic plaque formation. Sci. Rep. 2020, 10, 6127. [Google Scholar] [CrossRef]
- Wang, X.; Zheng, Y.; Ma, Y.; Du, L.; Chu, F.; Gu, H.; Dahlgren, R.A.; Li, Y.; Wang, H. Lipid metabolism disorder induced by up-regulation of miR-125b and miR-144 following OI-diketone antibiotic exposure to F0-zebrafish (Danio rerio). Ecotoxicol. Environ. Saf. 2018, 164, 243–252. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.M.; Liu, X.X.; Wei, G.Q.; Da, Y.N.; Cha, L.; Ma, C.S. Prediction of long-term outcome after acute myocardial infarction using circulating miR-145. Scand. J. Clin. Lab. Investig. 2015, 75, 85–91. [Google Scholar] [CrossRef]
- Zhang, M.; Cheng, Y.J.; Sara, J.D.; Liu, L.J.; Liu, L.P.; Zhao, X.; Gao, H. Circulating MicroRNA-145 is Associated with Acute Myocardial Infarction and Heart Failure. Chin. Med. J. 2017, 130, 51–56. [Google Scholar] [CrossRef]
- Du, Y.; Yang, S.H.; Li, S.; Cui, C.J.; Zhang, Y.; Zhu, C.G.; Guo, Y.L.; Wu, N.Q.; Gao, Y.; Sun, J.; et al. Circulating MicroRNAs as Novel Diagnostic Biomarkers for Very Early-onset (40 years) Coronary Artery Disease. Biomed. Environ. Sci. BES 2016, 29, 545–554. [Google Scholar]
- Gao, H.; Guddeti, R.R.; Matsuzawa, Y.; Liu, L.P.; Su, L.X.; Guo, D.; Nie, S.P.; Du, J.; Zhang, M. Plasma Levels of microRNA-145 Are Associated with Severity of Coronary Artery Disease. PLoS ONE 2015, 10, e0123477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Cheng, H.; Yue, Y.; Li, S.; Zhang, D.; He, R. H19 knockdown suppresses proliferation and induces apoptosis by regulating miR-148b/WNT/β-catenin in ox-LDL -stimulated vascular smooth muscle cells. J. Biomed. Sci. 2018, 25, 11. [Google Scholar] [CrossRef] [Green Version]
- Goedeke, L.; Rotllan, N.; Canfran-Duque, A.; Aranda, J.F.; Ramirez, C.M.; Araldi, E.; Lin, C.S.; Anderson, N.N.; Wagschal, A.; de Cabo, R.; et al. MicroRNA-148a regulates LDL receptor and ABCA1 expression to control circulating lipoprotein levels. Nat. Med. 2015, 21, 1280–1289. [Google Scholar] [CrossRef] [Green Version]
- Tang, X.E.; Li, H.; Chen, L.Y.; Xia, X.D.; Zhao, Z.W.; Zheng, X.L.; Zhao, G.J.; Tang, C.K. IL-8 negatively regulates ABCA1 expression and cholesterol efflux via upregulating miR-183 in THP-1 macrophage-derived foam cells. Cytokine 2019, 122, 154385. [Google Scholar] [CrossRef]
- Li, S.H.; Su, S.Y.; Liu, J.L. Differential Regulation of microRNAs in Patients with Ischemic Stroke. Curr. Neurovascular Res. 2015, 12, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.F.; Yang, Y.; Yang, X.Y.; Tong, Q. MiR-188-3p upregulation results in the inhibition of macrophage proinflammatory activities and atherosclerosis in ApoE-deficient mice. Thromb. Res. 2018, 171, 55–61. [Google Scholar] [CrossRef]
- Miao, H.; Zeng, H.; Gong, H. microRNA-212 promotes lipid accumulation and attenuates cholesterol efflux in THP-1 human macrophages by targeting SIRT1. Gene 2018, 643, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Shan, Z.; Qin, S.; Li, W.; Wu, W.; Yang, J.; Chu, M.; Li, X.; Huo, Y.; Schaer, G.L.; Wang, S.; et al. An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells: Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis. J. Am. Coll. Cardiol. 2015, 65, 2526–2537. [Google Scholar] [CrossRef] [Green Version]
- Wu, W.; Shan, Z.; Wang, R.; Chang, G.; Wang, M.; Wu, R.; Li, Z.; Zhang, C.; Li, W.; Wang, S. Overexpression of miR-223 inhibits foam cell formation by inducing autophagy in vascular smooth muscle cells. Am. J. Transl. Res. 2019, 11, 4326–4336. [Google Scholar]
- Singh, S.; de Ronde, M.W.J.; Kok, M.G.M.; Beijk, M.A.; De Winter, R.J.; van der Wal, A.C.; Sondermeijer, B.M.; Meijers, J.C.M.; Creemers, E.E.; Pinto-Sietsma, S.J. MiR-223-3p and miR-122-5p as circulating biomarkers for plaque instability. Open Heart 2020, 7, e001223. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ling, S.; Sun, W.; Liu, T.; Li, Y.; Zhong, G.; Zhao, D.; Zhang, P.; Song, J.; Jin, X.; et al. Circulating microRNAs correlated with the level of coronary artery calcification in symptomatic patients. Sci. Rep. 2015, 5, 16099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, B.J.; Nie, X.M.; Zeng, X.L.; Yuan, H.; Ma, X.; Zhao, Y.X.; Zhou, Y.J. [Expression of platelet miR-223 in coronary artery disease patients and its clinical significance]. Zhonghua Yi Xue Za Zhi 2018, 98, 1766–1770. [Google Scholar] [PubMed]
- Vickers, K.C.; Palmisano, B.T.; Shoucri, B.M.; Shamburek, R.D.; Remaley, A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011, 13, 423–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.F.; Zhang, Y.; Zheng, Q.X.; Zhang, Y.; Zhou, H.H.; Cui, L.M. Association between elevated plasma microRNA-223 content and severity of coronary heart disease. Scand. J. Clin. Lab. Investig. 2018, 78, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Saadatian, Z.; Nariman-Saleh-Fam, Z.; Bastami, M.; Mansoori, Y.; Khaheshi, I.; Parsa, S.A.; Daraei, A.; Vahed, S.Z.; Yousefi, B.; Kafil, H.S.; et al. Dysregulated expression of STAT1, miR-150, and miR-223 in peripheral blood mononuclear cells of coronary artery disease patients with significant or insignificant stenosis. J. Cell. Biochem. 2019, 120, 19810–19824. [Google Scholar] [CrossRef]
- Vickers, K.C.; Landstreet, S.R.; Levin, M.G.; Shoucri, B.M.; Toth, C.L.; Taylor, R.C.; Palmisano, B.T.; Tabet, F.; Cui, H.L.; Rye, K.A.; et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef] [Green Version]
- Meiler, S.; Baumer, Y.; Toulmin, E.; Seng, K.; Boisvert, W.A. MicroRNA 302a is a novel modulator of cholesterol homeostasis and atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 323–331. [Google Scholar] [CrossRef] [Green Version]
- Hoekstra, M.; van der Sluis, R.J.; Kuiper, J.; Van Berkel, T.J. Nonalcoholic fatty liver disease is associated with an altered hepatocyte microRNA profile in LDL receptor knockout mice. J. Nutr. Biochem. 2012, 23, 622–628. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Li, C.; Zhang, Y.; Zhou, X.; Liu, Y.; Lu, C. LncRNA MEG3-derived miR-361-5p regulate vascular smooth muscle cells proliferation and apoptosis by targeting ABCA1. Am. J. Transl. Res. 2019, 11, 3600–3609. [Google Scholar] [PubMed]
- Wang, D.; Yan, X.; Xia, M.; Yang, Y.; Li, D.; Li, X.; Song, F.; Ling, W. Coenzyme Q10 promotes macrophage cholesterol efflux by regulation of the activator protein-1/miR-378/ATP-binding cassette transporter G1-signaling pathway. Arterioscler. Thromb. Vasc. Biol. 2014, 34, 1860–1870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Gao, F.; Wang, X.; Wu, J.; Lu, K.; Liu, M.; Li, R.; Ding, L.; Wang, R. Circulating microRNA-378 levels serve as a novel biomarker for assessing the severity of coronary stenosis in patients with coronary artery disease. Biosci. Rep. 2019, 39. [Google Scholar] [CrossRef] [Green Version]
- Prats-Puig, A.; Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Bonet, N.; Ricart, W.; Lopez-Bermejo, A.; Fernandez-Real, J.M. Changes in circulating microRNAs are associated with childhood obesity. J. Clin. Endocrinol. Metab. 2013, 98, E1655–E1660. [Google Scholar] [CrossRef]
- Santamaria-Martos, F.; Benitez, I.; Pinilla, L.; Ortega, F.; Zapater, A.; Giron, C.; Minguez, O.; Gomez, S.; Vaca, R.; Fernandez-Real, J.M.; et al. MicroRNA Profile of Cardiovascular Risk in Patients with Obstructive Sleep Apnea. Respir. Int. Rev. Thorac. Dis. 2020, 99, 1122–1128. [Google Scholar] [CrossRef]
- Liu, D.; Zhang, M.; Xie, W.; Lan, G.; Cheng, H.P.; Gong, D.; Huang, C.; Lv, Y.C.; Yao, F.; Tan, Y.L.; et al. MiR-486 regulates cholesterol efflux by targeting HAT1. Biochem. Biophys. Res. Commun. 2016, 472, 418–424. [Google Scholar] [CrossRef]
- Zhao, R.; Feng, J.; He, G. miR-613 regulates cholesterol efflux by targeting LXRα and ABCA1 in PPARγ activated THP-1 macrophages. Biochem. Biophys. Res. Commun. 2014, 448, 329–334. [Google Scholar] [CrossRef]
- Ramirez, C.M.; Davalos, A.; Goedeke, L.; Salerno, A.G.; Warrier, N.; Cirera-Salinas, D.; Suarez, Y.; Fernandez-Hernando, C. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler. Thromb. Vasc. Biol. 2011, 31, 2707–2714. [Google Scholar] [CrossRef] [Green Version]
- Mandolini, C.; Santovito, D.; Marcantonio, P.; Buttitta, F.; Bucci, M.; Ucchino, S.; Mezzetti, A.; Cipollone, F. Identification of microRNAs 758 and 33b as potential modulators of ABCA1 expression in human atherosclerotic plaques. Nutr. Metab. Cardiovasc. Dis. NMCD 2015, 25, 202–209. [Google Scholar] [CrossRef]
- O’Neill, S.; Larsen, M.B.; Gregersen, S.; Hermansen, K.; O’Driscoll, L. miR-758-3p: A blood-based biomarker thats influence on the expression of CERP/ABCA1 may contribute to the progression of obesity to metabolic syndrome. Oncotarget 2018, 9, 9379–9390. [Google Scholar] [CrossRef] [Green Version]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Naar, A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef] [Green Version]
- Rayner, K.J.; Suarez, Y.; Davalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernandez-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Yoon, H.; Horie, T.; Burchett, J.M.; Restivo, J.L.; Rotllan, N.; Ramirez, C.M.; Verghese, P.B.; Ihara, M.; Hoe, H.S.; et al. microRNA-33 Regulates ApoE Lipidation and Amyloid-β Metabolism in the Brain. J. Neurosci. 2015, 35, 14717–14726. [Google Scholar] [CrossRef]
- Zhao, L.; Huang, J.; Zhu, Y.; Han, S.; Qing, K.; Wang, J.; Feng, Y. miR-33-5p knockdown attenuates abdominal aortic aneurysm progression via promoting target adenosine triphosphate-binding cassette transporter A1 expression and activating the PI3K/Akt signaling pathway. Perfusion 2020, 35, 57–65. [Google Scholar] [CrossRef] [PubMed]
- Rayner, K.J.; Sheedy, F.J.; Esau, C.C.; Hussain, F.N.; Temel, R.E.; Parathath, S.; van Gils, J.M.; Rayner, A.J.; Chang, A.N.; Suarez, Y.; et al. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J. Clin. Investig. 2011, 121, 2921–2931. [Google Scholar] [CrossRef] [Green Version]
- Price, N.L.; Rotllan, N.; Zhang, X.; Canfran-Duque, A.; Nottoli, T.; Suarez, Y.; Fernandez-Hernando, C. Specific Disruption of Abca1 Targeting Largely Mimics the Effects of miR-33 Knockout on Macrophage Cholesterol Efflux and Atherosclerotic Plaque Development. Circ. Res. 2019, 124, 874–880. [Google Scholar] [CrossRef] [PubMed]
- Mao, M.; Lei, H.; Liu, Q.; Chen, Y.; Zhao, L.; Li, Q.; Luo, S.; Zuo, Z.; He, Q.; Huang, W.; et al. Effects of miR-33a-5P on ABCA1/G1-mediated cholesterol efflux under inflammatory stress in THP-1 macrophages. PLoS ONE 2014, 9, e109722. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.H.; Kim, G.J.; Umemura, T.; Lee, S.G.; Cho, K.J. Aberrant expression of plasma microRNA-33a in an atherosclerosis-risk group. Mol. Biol. Rep. 2017, 44, 79–88. [Google Scholar] [CrossRef] [Green Version]
- Reddy, L.L.; Shah, S.A.V.; Ponde, C.K.; Rajani, R.M.; Ashavaid, T.F. Circulating miRNA-33: A potential biomarker in patients with coronary artery disease. Biomarker 2019, 24, 36–42. [Google Scholar] [CrossRef] [PubMed]
- Nishino, T.; Horie, T.; Baba, O.; Sowa, N.; Hanada, R.; Kuwabara, Y.; Nakao, T.; Nishiga, M.; Nishi, H.; Nakashima, Y.; et al. SREBF1/MicroRNA-33b Axis Exhibits Potent Effect on Unstable Atherosclerotic Plaque Formation In Vivo. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2460–2473. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Baba, O.; Kuwabara, Y.; Chujo, Y.; Watanabe, S.; Kinoshita, M.; Horiguchi, M.; Nakamura, T.; Chonabayashi, K.; Hishizawa, M.; et al. MicroRNA-33 deficiency reduces the progression of atherosclerotic plaque in ApoE-/- mice. J. Am. Heart Assoc. 2012, 1, e003376. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Ono, K.; Horiguchi, M.; Nishi, H.; Nakamura, T.; Nagao, K.; Kinoshita, M.; Kuwabara, Y.; Marusawa, H.; Iwanaga, Y.; et al. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 17321–17326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama, S.; Horie, T.; Nishino, T.; Baba, O.; Sowa, N.; Miyasaka, Y.; Kuwabara, Y.; Nakao, T.; Nishiga, M.; Nishi, H.; et al. Identification of Differential Roles of MicroRNA-33a and -33b During Atherosclerosis Progression with Genetically Modified Mice. J. Am. Heart Assoc. 2019, 8, e012609. [Google Scholar] [CrossRef]
- Rotllan, N.; Ramirez, C.M.; Aryal, B.; Esau, C.C.; Fernandez-Hernando, C. Therapeutic silencing of microRNA-33 inhibits the progression of atherosclerosis in Ldlr-/- mice—Brief report. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 1973–1977. [Google Scholar] [CrossRef] [Green Version]
- Ouimet, M.; Ediriweera, H.N.; Gundra, U.M.; Sheedy, F.J.; Ramkhelawon, B.; Hutchison, S.B.; Rinehold, K.; van Solingen, C.; Fullerton, M.D.; Cecchini, K.; et al. MicroRNA-33-dependent regulation of macrophage metabolism directs immune cell polarization in atherosclerosis. J. Clin. Investig. 2015, 125, 4334–4348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouimet, M.; Ediriweera, H.; Afonso, M.S.; Ramkhelawon, B.; Singaravelu, R.; Liao, X.; Bandler, R.C.; Rahman, K.; Fisher, E.A.; Rayner, K.J.; et al. microRNA-33 Regulates Macrophage Autophagy in Atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 1058–1067. [Google Scholar] [CrossRef] [Green Version]
- Price, N.L.; Rotllan, N.; Canfran-Duque, A.; Zhang, X.; Pati, P.; Arias, N.; Moen, J.; Mayr, M.; Ford, D.A.; Baldan, T.; et al. Genetic Dissection of the Impact of miR-33a and miR-33b during the Progression of Atherosclerosis. Cell Rep. 2017, 21, 1317–1330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [Green Version]
- Arslan, S.; Berkan, O.; Lalem, T.; Ozbilum, N.; Goksel, S.; Korkmaz, O.; Cetin, N.; Devaux, Y. Long non-coding RNAs in the atherosclerotic plaque. Atherosclerosis 2017, 266, 176–181. [Google Scholar] [CrossRef]
- Liu, L.; Tan, L.; Yao, J.; Yang, L. Long non-coding RNA MALAT1 regulates cholesterol accumulation in ox-LDL-induced macrophages via the microRNA-17-5p/ABCA1 axis. Mol. Med. Rep. 2020, 21, 1761–1770. [Google Scholar] [CrossRef] [Green Version]
- Hennessy, E.J.; van Solingen, C.; Scacalossi, K.R.; Ouimet, M.; Afonso, M.S.; Prins, J.; Koelwyn, G.J.; Sharma, M.; Ramkhelawon, B.; Carpenter, S.; et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat. Metab. 2019, 1, 98–110. [Google Scholar] [CrossRef]
- Diao, L.; Bai, L.; Jiang, X.; Li, J.; Zhang, Q. Long-chain noncoding RNA GAS5 mediates oxidative stress in cardiac microvascular endothelial cells injury. J. Cell. Physiol. 2019, 234, 17649–17662. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.W.; Yang, J.Y.; Ma, X.; Chen, Z.P.; Hu, Y.R.; Zhao, J.Y.; Li, S.F.; Qiu, Y.R.; Lu, J.B.; Wang, Y.C.; et al. A lincRNA-DYNLRB2-2/GPR119/GLP-1R/ABCA1-dependent signal transduction pathway is essential for the regulation of cholesterol homeostasis. J. Lipid Res. 2014, 55, 681–697. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Shen, S.; Ding, S.; Wang, L. LincRNA DYN-LRB2-2 upregulates cholesterol efflux by decreasing TLR2 expression in macrophages. J. Cell. Biochem. 2018, 119, 1911–1921. [Google Scholar] [CrossRef]
- Lan, X.; Yan, J.; Ren, J.; Zhong, B.; Li, J.; Li, Y.; Liu, L.; Yi, J.; Sun, Q.; Yang, X.; et al. A novel long noncoding RNA Lnc-HC binds hnRNPA2B1 to regulate expressions of Cyp7a1 and Abca1 in hepatocytic cholesterol metabolism. Hepatology 2016, 64, 58–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ou, M.; Li, X.; Zhao, S.; Cui, S.; Tu, J. Long non-coding RNA CDKN2B-AS1 contributes to atherosclerotic plaque formation by forming RNA-DNA triplex in the CDKN2B promoter. EBioMedicine 2020, 55, 102694. [Google Scholar] [CrossRef]
- Yang, L.; Li, T. LncRNA TUG1 regulates ApoM to promote atherosclerosis progression through miR-92a/FXR1 axis. J. Cell. Mol. Med. 2020, 24, 8836–8848. [Google Scholar] [CrossRef] [PubMed]
- Filippenkov, I.B.; Kalinichenko, E.O.; Limborska, S.A.; Dergunova, L.V. Circular RNAs-one of the enigmas of the brain. Neurogenetics 2017, 18, 1–6. [Google Scholar] [CrossRef]
- Filippenkov, I.B.; Sudarkina, O.Y.; Limborska, S.A.; Dergunova, L.V. Circular RNA of the human sphingomyelin synthase 1 gene: Multiple splice variants, evolutionary conservatism and expression in different tissues. RNA Biol. 2015, 12, 1030–1042. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef] [PubMed]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef] [Green Version]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Shen, L.; Chen, H.; Wang, R.; Zang, T.; Qian, J.; Ge, J. circDENND1B Participates in the Antiatherosclerotic Effect of IL-1β Monoclonal Antibody in Mouse by Promoting Cholesterol Efflux via miR-17-5p/Abca1 Axis. Front. Cell Dev. Biol. 2021, 9, 652032. [Google Scholar] [CrossRef]
- Klein, I.; Sarkadi, B.; Varadi, A. An inventory of the human ABC proteins. Biochim. Biophys. Acta 1999, 1461, 237–262. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, M.A.; Venkateswaran, A.; Tarr, P.T.; Xenarios, I.; Kudoh, J.; Shimizu, N.; Edwards, P.A. Characterization of the human ABCG1 gene: Liver X receptor activates an internal promoter that produces a novel transcript encoding an alternative form of the protein. J. Biol. Chem. 2001, 276, 39438–39447. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Lan, D.; Chen, W.; Matsuura, F.; Tall, A.R. ATP-binding cassette transporters G1 and G4 mediate cellular cholesterol efflux to high-density lipoproteins. Proc. Natl. Acad. Sci. USA 2004, 101, 9774–9779. [Google Scholar] [CrossRef] [Green Version]
- Miroshnikova, V.V.; Demina, E.P.; Maiorov, N.V.; Davydenko, V.V.; Kur’ianov, P.S.; Vavilov, V.N.; Vinogradov, A.G.; Denisenko, A.D.; Shvartsman, A.L. ABCG1 transporter gene expression in peripheral blood mononuclear cells of patients with atherosclerosis. Tsitologiia 2014, 56, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Mauldin, J.P.; Nagelin, M.H.; Wojcik, A.J.; Srinivasan, S.; Skaflen, M.D.; Ayers, C.R.; McNamara, C.A.; Hedrick, C.C. Reduced expression of ATP-binding cassette transporter G1 increases cholesterol accumulation in macrophages of patients with type 2 diabetes mellitus. Circulation 2008, 117, 2785–2792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kennedy, M.A.; Barrera, G.C.; Nakamura, K.; Baldan, A.; Tarr, P.; Fishbein, M.C.; Frank, J.; Francone, O.L.; Edwards, P.A. ABCG1 has a critical role in mediating cholesterol efflux to HDL and preventing cellular lipid accumulation. Cell Metab. 2005, 1, 121–131. [Google Scholar] [CrossRef] [Green Version]
- Gelissen, I.C.; Harris, M.; Rye, K.A.; Quinn, C.; Brown, A.J.; Kockx, M.; Cartland, S.; Packianathan, M.; Kritharides, L.; Jessup, W. ABCA1 and ABCG1 synergize to mediate cholesterol export to apoA-I. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 534–540. [Google Scholar] [CrossRef] [Green Version]
- Meurs, I.; Lammers, B.; Zhao, Y.; Out, R.; Hildebrand, R.B.; Hoekstra, M.; Van Berkel, T.J.; Van, E.M. The effect of ABCG1 deficiency on atherosclerotic lesion development in LDL receptor knockout mice depends on the stage of atherogenesis. Atherosclerosis 2012, 221, 41–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ranalletta, M.; Wang, N.; Han, S.; Yvan-Charvet, L.; Welch, C.; Tall, A.R. Decreased atherosclerosis in low-density lipoprotein receptor knockout mice transplanted with Abcg1-/- bone marrow. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 2308–2315. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.Q.; Yan, X.W.; Li, P.T.; Ji, X.H. Analysis of differential gene expression by RNA-seq data in ABCG1 knockout mice. Gene 2019, 689, 24–33. [Google Scholar] [CrossRef]
- Schou, J.; Frikke-Schmidt, R.; Kardassis, D.; Thymiakou, E.; Nordestgaard, B.G.; Jensen, G.; Grande, P.; Tybjaerg-Hansen, A. Genetic variation in ABCG1 and risk of myocardial infarction and ischemic heart disease. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 506–515. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Wang, W.; Zhang, L.; Qi, L.P.; Li, L.Y.; Chen, L.F.; Fang, Q.; Dang, A.M.; Yan, X.W. A polymorphism in the ABCG1 promoter is functionally associated with coronary artery disease in a Chinese Han population. Atherosclerosis 2011, 219, 648–654. [Google Scholar] [CrossRef]
- Furuyama, S.; Uehara, Y.; Zhang, B.; Baba, Y.; Abe, S.; Iwamoto, T.; Miura, S.; Saku, K. Genotypic Effect of ABCG1 gene promoter -257T>G polymorphism on coronary artery disease severity in Japanese men. J. Atheroscler. Thromb. 2009, 16, 194–200. [Google Scholar] [CrossRef] [Green Version]
- Takata, K.; Honda, S.; Sidharta, S.L.; Duong, M.; Shishikura, D.; Kim, S.W.; Andrews, J.; Di Bartolo, B.A.; Psaltis, P.J.; Bursill, C.A.; et al. Associations of ABCG1-mediated cholesterol efflux capacity with coronary artery lipid content assessed by near-infrared spectroscopy. Cardiovasc. Diagn. Ther. 2019, 9, 310–318. [Google Scholar] [CrossRef] [PubMed]
- Peng, P.; Wang, L.; Yang, X.; Huang, X.; Ba, Y.; Chen, X.; Guo, J.; Lian, J.; Zhou, J. A preliminary study of the relationship between promoter methylation of the ABCG1, GALNT2 and HMGCR genes and coronary heart disease. PLoS ONE 2014, 9, e102265. [Google Scholar] [CrossRef] [Green Version]
- Hedman, A.K.; Mendelson, M.M.; Marioni, R.E.; Gustafsson, S.; Joehanes, R.; Irvin, M.R.; Zhi, D.; Sandling, J.K.; Yao, C.; Liu, C.; et al. Epigenetic Patterns in Blood Associated with Lipid Traits Predict Incident Coronary Heart Disease Events and Are Enriched for Results From Genome-Wide Association Studies. Circ. Cardiovasc. Genet. 2017, 10, e001487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, X.; Li, J.; Wu, T.; Wu, Y.; Tang, X.; Gao, P.; Li, L.; Wang, M.; Wu, Y.; Wang, X.; et al. Overall and sex-specific associations between methylation of the ABCG1 and APOE genes and ischemic stroke or other atherosclerosis-related traits in a sibling study of Chinese population. Clin. Epigenetics 2019, 11, 189. [Google Scholar] [CrossRef]
- Pfeiffer, L.; Wahl, S.; Pilling, L.C.; Reischl, E.; Sandling, J.K.; Kunze, S.; Holdt, L.M.; Kretschmer, A.; Schramm, K.; Adamski, J.; et al. DNA methylation of lipid-related genes affects blood lipid levels. Circ. Cardiovasc. Genet. 2015, 8, 334–342. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Cai, Q.; Zhang, D.; Fan, J.; Hu, S.; Venners, S.A. Effect of ABCG1 gene DNA methylations on the lipid-lowering efficacy of simvastatin. Pharmacogenomics 2021, 22, 27–39. [Google Scholar] [CrossRef]
- Liu, J.; Huan, C.; Chakraborty, M.; Zhang, H.; Lu, D.; Kuo, M.S.; Cao, G.; Jiang, X.C. Macrophage sphingomyelin synthase 2 deficiency decreases atherosclerosis in mice. Circ. Res. 2009, 105, 295–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sano, O.; Kobayashi, A.; Nagao, K.; Kumagai, K.; Kioka, N.; Hanada, K.; Ueda, K.; Matsuo, M. Sphingomyelin-dependence of cholesterol efflux mediated by ABCG1. J. Lipid Res. 2007, 48, 2377–2384. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Ranalletta, M.; Matsuura, F.; Peng, F.; Tall, A.R. LXR-induced redistribution of ABCG1 to plasma membrane in macrophages enhances cholesterol mass efflux to HDL. Arterioscler. Thromb. Vasc. Biol. 2006, 26, 1310–1316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabol, S.L.; Brewer, H.B., Jr.; Santamarina-Fojo, S. The human ABCG1 gene: Identification of LXR response elements that modulate expression in macrophages and liver. J. Lipid Res. 2005, 46, 2151–2167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorkowski, S.; Rust, S.; Engel, T.; Jung, E.; Tegelkamp, K.; Galinski, E.A.; Assmann, G.; Cullen, P. Genomic sequence and structure of the human ABCG1 (ABC8) gene. Biochem. Biophys. Res. Commun. 2001, 280, 121–131. [Google Scholar] [CrossRef]
- Xu, B.M.; Xiao, L.; Kang, C.M.; Ding, L.; Guo, F.X.; Li, P.; Lu, Z.F.; Wu, Q.; Xu, Y.J.; Bai, H.L.; et al. LncRNA AC096664.3/PPAR-Oi/ABCG1-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis. J. Cell. Biochem. 2019, 120, 13775–13782. [Google Scholar] [CrossRef]
- Cai, C.; Zhu, H.; Ning, X.; Li, L.; Yang, B.; Chen, S.; Wang, L.; Lu, X.; Gu, D. LncRNA ENST00000602558.1 regulates ABCG1 expression and cholesterol efflux from vascular smooth muscle cells through a p65-dependent pathway. Atherosclerosis 2019, 285, 31–39. [Google Scholar] [CrossRef]
- Li, X.; Ji, Z.; Li, S.; Sun, Y.N.; Liu, J.; Liu, Y.; Tian, W.; Zhou, Y.T.; Shang, X.M. miR-146a-5p Antagonized AGEs- and P.g-LPS-Induced ABCA1 and ABCG1 Dysregulation in Macrophages via IRAK-1 Downregulation. Inflammation 2015, 38, 1761–1768. [Google Scholar] [CrossRef]
- Li, H.N.; Zhao, X.; Zha, Y.J.; Du, F.; Liu, J.; Sun, L. miR-146a-5p suppresses ATP-binding cassette subfamily G member 1 dysregulation in patients with refractory Mycoplasma pneumoniae via interleukin 1 receptor-associated kinase 1 downregulation. Int. J. Mol. Med. 2019, 44, 2003–2014. [Google Scholar] [CrossRef] [Green Version]
- Marquart, T.J.; Allen, R.M.; Ory, D.S.; Baldan, A. miR-33 links SREBP-2 induction to repression of sterol transporters. Proc. Natl. Acad. Sci. USA 2010, 107, 12228–12232. [Google Scholar] [CrossRef] [Green Version]
- Horie, T.; Nishino, T.; Baba, O.; Kuwabara, Y.; Nakao, T.; Nishiga, M.; Usami, S.; Izuhara, M.; Nakazeki, F.; Ide, Y.; et al. MicroRNA-33b knock-in mice for an intron of sterol regulatory element-binding factor 1 (Srebf1) exhibit reduced HDL-C in vivo. Sci. Rep. 2014, 4, 5312. [Google Scholar] [CrossRef] [Green Version]
- Hussain, M.M.; Goldberg, I.J. Human MicroRNA-33b Promotes Atherosclerosis in Apoe(-/-) Mice. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 2272–2275. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.P.; Amar, M.J.; Vaisman, B.; Bocharov, A.V.; Vishnyakova, T.G.; Freeman, L.A.; Kurlander, R.J.; Patterson, A.P.; Becker, L.C.; Remaley, A.T. Scavenger receptor-BI is a receptor for lipoprotein(a). J. Lipid Res. 2013, 54, 2450–2457. [Google Scholar] [CrossRef] [Green Version]
- Shchelkunova, T.A.; Morozov, I.A.; Rubtsov, P.M.; Samokhodskaya, L.M.; Andrianova, I.V.; Sobenin, I.A.; Orekhov, A.N.; Smirnov, A.N. Changes in levels of gene expression in human aortal intima during atherogenesis. Biochem. Biokhimiia 2013, 78, 463–470. [Google Scholar] [CrossRef]
- Dergunov, A.D.; Litvinov, D.Y.; Bazaeva, E.V.; Dmitrieva, V.G.; Nosova, E.V.; Rozhkova, A.V.; Dergunova, L.V. Relation of High-Density Lipoprotein Charge Heterogeneity, Cholesterol Efflux Capacity, and the Expression of High-Density Lipoprotein-Related Genes in Mononuclear Cells to the HDL-Cholesterol Level. Lipids 2018, 53, 979–991. [Google Scholar] [CrossRef]
- Dergunova, L.V.; Nosova, E.V.; Dmitrieva, V.G.; Rozhkova, A.V.; Bazaeva, E.V.; Limborska, S.A.; Dergunov, A.D. HDL cholesterol is associated with PBMC expression of genes involved in HDL metabolism and atherogenesis. J. Med. Biochem. 2020, 39, 372–383. [Google Scholar] [CrossRef] [Green Version]
- Van Eck, M.; Twisk, J.; Hoekstra, M.; Van Rij, B.T.; Van der Lans, C.A.; Bos, I.S.; Kruijt, J.K.; Kuipers, F.; Van Berkel, T.J. Differential effects of scavenger receptor BI deficiency on lipid metabolism in cells of the arterial wall and in the liver. J. Biol. Chem. 2003, 278, 23699–23705. [Google Scholar] [CrossRef] [Green Version]
- Braun, A.; Trigatti, B.L.; Post, M.J.; Sato, K.; Simons, M.; Edelberg, J.M.; Rosenberg, R.D.; Schrenzel, M.; Krieger, M. Loss of SR-BI expression leads to the early onset of occlusive atherosclerotic coronary artery disease, spontaneous myocardial infarctions, severe cardiac dysfunction, and premature death in apolipoprotein E-deficient mice. Circ. Res. 2002, 90, 270–276. [Google Scholar] [CrossRef]
- Zhang, W.; Yancey, P.G.; Su, Y.R.; Babaev, V.R.; Zhang, Y.; Fazio, S.; Linton, M.F. Inactivation of macrophage scavenger receptor class B type I promotes atherosclerotic lesion development in apolipoprotein E-deficient mice. Circulation 2003, 108, 2258–2263. [Google Scholar] [CrossRef] [Green Version]
- Covey, S.D.; Krieger, M.; Wang, W.; Penman, M.; Trigatti, B.L. Scavenger receptor class B type I-mediated protection against atherosclerosis in LDL receptor-negative mice involves its expression in bone marrow-derived cells. Arterioscler. Thromb. Vasc. Biol. 2003, 23, 1589–1594. [Google Scholar] [CrossRef] [Green Version]
- Tao, H.; Yancey, P.G.; Babaev, V.R.; Blakemore, J.L.; Zhang, Y.; Ding, L.; Fazio, S.; Linton, M.F. Macrophage SR-BI mediates efferocytosis via Src/PI3K/Rac1 signaling and reduces atherosclerotic lesion necrosis. J. Lipid Res. 2015, 56, 1449–1460. [Google Scholar] [CrossRef] [Green Version]
- Kozarsky, K.F.; Donahee, M.H.; Glick, J.M.; Krieger, M.; Rader, D.J. Gene transfer and hepatic overexpression of the HDL receptor SR-BI reduces atherosclerosis in the cholesterol-fed LDL receptor-deficient mouse. Arterioscler. Thromb. Vasc. Biol. 2000, 20, 721–727. [Google Scholar] [CrossRef] [Green Version]
- Huby, T.; Doucet, C.; Dachet, C.; Ouzilleau, B.; Ueda, Y.; Afzal, V.; Rubin, E.; Chapman, M.J.; Lesnik, P. Knockdown expression and hepatic deficiency reveal an atheroprotective role for SR-BI in liver and peripheral tissues. J. Clin. Investig. 2006, 116, 2767–2776. [Google Scholar] [CrossRef] [Green Version]
- Adorni, M.P.; Zimetti, F.; Billheimer, J.T.; Wang, N.; Rader, D.J.; Phillips, M.C.; Rothblat, G.H. The roles of different pathways in the release of cholesterol from macrophages. J. Lipid Res. 2007, 48, 2453–2462. [Google Scholar] [CrossRef] [Green Version]
- Yancey, P.G.; Jerome, W.G.; Yu, H.; Griffin, E.E.; Cox, B.E.; Babaev, V.R.; Fazio, S.; Linton, M.F. Severely altered cholesterol homeostasis in macrophages lacking apoE and SR-BI. J. Lipid Res. 2007, 48, 1140–1149. [Google Scholar] [CrossRef] [Green Version]
- Fadok, V.A.; Warner, M.L.; Bratton, D.L.; Henson, P.M. CD36 is required for phagocytosis of apoptotic cells by human macrophages that use either a phosphatidylserine receptor or the vitronectin receptor (alpha v beta 3). J. Immunol. 1998, 161, 6250–6257. [Google Scholar]
- Zanoni, P.; Khetarpal, S.A.; Larach, D.B.; Hancock-Cerutti, W.F.; Millar, J.S.; Cuchel, M.; DerOhannessian, S.; Kontush, A.; Surendran, P.; Saleheen, D.; et al. Rare variant in scavenger receptor BI raises HDL cholesterol and increases risk of coronary heart disease. Science 2016, 351, 1166–1171. [Google Scholar] [CrossRef] [Green Version]
- Samadi, S.; Farjami, Z.; Hosseini, Z.S.; Ferns, G.A.; Mohammadpour, A.H.; Tayefi, M.; Fal-Soleiman, H.; Moohebati, M.; Ghayour-Mobarhan, M.; Esmaily, H.; et al. Rare P376L variant in the SR-BI gene associates with HDL dysfunction and risk of cardiovascular disease. Clin. Biochem. 2019, 73, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Vergeer, M.; Korporaal, S.J.; Franssen, R.; Meurs, I.; Out, R.; Hovingh, G.K.; Hoekstra, M.; Sierts, J.A.; Dallinga-Thie, G.M.; Motazacker, M.M.; et al. Genetic variant of the scavenger receptor BI in humans. N. Engl. J. Med. 2011, 364, 136–145. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Sethi, A.; Yanek, L.R.; Knapper, C.; Nordestgaard, B.G.; Tybjaerg-Hansen, A.; Becker, D.M.; Mathias, R.A.; Remaley, A.T.; Becker, L.C. SCARB1 Gene Variants Are Associated with the Phenotype of Combined High High-Density Lipoprotein Cholesterol and High Lipoprotein (a). Circ. Cardiovasc. Genet. 2016, 9, 408–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manichaikul, A.; Wang, X.Q.; Musani, S.K.; Herrington, D.M.; Post, W.S.; Wilson, J.G.; Rich, S.S.; Rodriguez, A. Association of the Lipoprotein Receptor SCARB1 Common Missense Variant rs4238001 with Incident Coronary Heart Disease. PLoS ONE 2015, 10, e0125497. [Google Scholar] [CrossRef] [Green Version]
- Manichaikul, A.; Naj, A.C.; Herrington, D.; Post, W.; Rich, S.S.; Rodriguez, A. Association of SCARB1 variants with subclinical atherosclerosis and incident cardiovascular disease: The multi-ethnic study of atherosclerosis. Arterioscler. Thromb. Vasc. Biol. 2012, 32, 1991–1999. [Google Scholar] [CrossRef] [Green Version]
- Ritsch, A.; Sonderegger, G.; Sandhofer, A.; Stanzl, U.; Tancevski, I.; Eller, P.; Schgoer, W.; Wehinger, A.; Mueller, T.; Haltmayer, M.; et al. Scavenger receptor class B type I polymorphisms and peripheral arterial disease. Metab. Clin. Exp. 2007, 56, 1135–1141. [Google Scholar] [CrossRef] [PubMed]
- Naj, A.C.; West, M.; Rich, S.S.; Post, W.; Kao, W.H.; Wasserman, B.A.; Herrington, D.M.; Rodriguez, A. Association of scavenger receptor class B type I polymorphisms with subclinical atherosclerosis: The Multi-Ethnic Study of Atherosclerosis. Circ. Cardiovasc. Genet. 2010, 3, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Howson, J.M.M.; Zhao, W.; Barnes, D.R.; Ho, W.K.; Young, R.; Paul, D.S.; Waite, L.L.; Freitag, D.F.; Fauman, E.B.; Salfati, E.L.; et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat. Genet. 2017, 49, 1113–1119. [Google Scholar] [CrossRef] [Green Version]
- Webb, T.R.; Erdmann, J.; Stirrups, K.E.; Stitziel, N.O.; Masca, N.G.; Jansen, H.; Kanoni, S.; Nelson, C.P.; Ferrario, P.G.; Konig, I.R.; et al. Systematic Evaluation of Pleiotropy Identifies 6 Further Loci Associated with Coronary Artery Disease. J. Am. Coll. Cardiol. 2017, 69, 823–836. [Google Scholar] [CrossRef]
- Ma, R.; Zhu, X.; Yan, B. SCARB1 rs5888 gene polymorphisms in coronary heart disease: A systematic review and a meta-analysis. Gene 2018, 678, 280–287. [Google Scholar] [CrossRef]
- Helgadottir, A.; Sulem, P.; Thorgeirsson, G.; Gretarsdottir, S.; Thorleifsson, G.; Jensson, B.Ö.; Arnadottir, G.A.; Olafsson, I.; Eyjolfsson, G.I.; Sigurdardottir, O.; et al. Rare SCARB1 mutations associate with high-density lipoprotein cholesterol but not with coronary artery disease. Eur. Heart J. 2018, 39, 2172–2178. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Zhang, H.; Yang, A.; Ma, P.; Sun, L.; Deng, M.; Mao, C.; Xiong, J.; Sun, J.; Wang, N.; et al. Homocysteine accelerates atherosclerosis by inhibiting scavenger receptor class B member1 via DNMT3b/SP1 pathway. J. Mol. Cell. Cardiol. 2020, 138, 34–48. [Google Scholar] [CrossRef]
- Dong, B.; Singh, A.B.; Guo, G.L.; Young, M.; Liu, J. Activation of FXR by obeticholic acid induces hepatic gene expression of SR-BI through a novel mechanism of transcriptional synergy with the nuclear receptor LXR. Int. J. Mol. Med. 2019, 43, 1927–1938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malerod, L.; Sporstol, M.; Juvet, L.K.; Mousavi, A.; Gjoen, T.; Berg, T. Hepatic scavenger receptor class B, type I is stimulated by peroxisome proliferator-activated receptor gamma and hepatocyte nuclear factor 4alpha. Biochem. Biophys. Res. Commun. 2003, 305, 557–565. [Google Scholar] [CrossRef]
- Huangfu, N.; Xu, Z.; Zheng, W.; Wang, Y.; Cheng, J.; Chen, X. LncRNA MALAT1 regulates oxLDL-induced CD36 expression via activating β-catenin. Biochem. Biophys. Res. Commun. 2018, 495, 2111–2117. [Google Scholar] [CrossRef]
- Wang, S.; Sun, Z.; Zhang, X.; Li, Z.; Wu, M.; Zhao, W.; Wang, H.; Chen, T.; Yan, H.; Zhu, J. Wnt1 positively regulates CD36 expression via TCF4 and PPAR-γ in macrophages. Cell. Physiol. Biochem. 2015, 35, 1289–1302. [Google Scholar] [CrossRef]
- Nicholls, S.J.; Schwartz, G.G.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Sweeney, M.; et al. Apabetalone and hospitalization for heart failure in patients following an acute coronary syndrome: A prespecified analysis of the BETonMACE study. Cardiovasc. Diabetol. 2021, 20, 13. [Google Scholar] [CrossRef]
- Jahagirdar, R.; Zhang, H.; Azhar, S.; Tobin, J.; Attwell, S.; Yu, R.; Wu, J.; McLure, K.G.; Hansen, H.C.; Wagner, G.S.; et al. A novel BET bromodomain inhibitor, RVX-208, shows reduction of atherosclerosis in hyperlipidemic ApoE deficient mice. Atherosclerosis 2014, 236, 91–100. [Google Scholar] [CrossRef]
- Bailey, D.; Jahagirdar, R.; Gordon, A.; Hafiane, A.; Campbell, S.; Chatur, S.; Wagner, G.S.; Hansen, H.C.; Chiacchia, F.S.; Johansson, J.; et al. RVX-208: A small molecule that increases apolipoprotein A-I and high-density lipoprotein cholesterol in vitro and in vivo. J. Am. Coll. Cardiol. 2010, 55, 2580–2589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsujikawa, L.M.; Fu, L.; Das, S.; Halliday, C.; Rakai, B.D.; Stotz, S.C.; Sarsons, C.D.; Gilham, D.; Daze, E.; Wasiak, S.; et al. Apabetalone (RVX-208) reduces vascular inflammation in vitro and in CVD patients by a BET-dependent epigenetic mechanism. Clin. Epigenetics 2019, 11, 102. [Google Scholar] [CrossRef]
- Shishikura, D.; Kataoka, Y.; Honda, S.; Takata, K.; Kim, S.W.; Andrews, J.; Psaltis, P.J.; Sweeney, M.; Kulikowski, E.; Johansson, J.; et al. The Effect of Bromodomain and Extra-Terminal Inhibitor Apabetalone on Attenuated Coronary Atherosclerotic Plaque: Insights from the ASSURE Trial. Am. J. Cardiovasc. Drugs 2019, 19, 49–57. [Google Scholar] [CrossRef]
- Ray, K.K.; Nicholls, S.J.; Buhr, K.A.; Ginsberg, H.N.; Johansson, J.O.; Kalantar-Zadeh, K.; Kulikowski, E.; Toth, P.P.; Wong, N.; Sweeney, M.; et al. Effect of Apabetalone Added to Standard Therapy on Major Adverse Cardiovascular Events in Patients with Recent Acute Coronary Syndrome and Type 2 Diabetes: A Randomized Clinical Trial. JAMA 2020, 323, 1565–1573. [Google Scholar] [CrossRef]
- Nicholls, S.J.; Puri, R.; Wolski, K.; Ballantyne, C.M.; Barter, P.J.; Brewer, H.B.; Kastelein, J.J.; Hu, B.; Uno, K.; Kataoka, Y.; et al. Effect of the BET Protein Inhibitor, RVX-208, on Progression of Coronary Atherosclerosis: Results of the Phase 2b, Randomized, Double-Blind, Multicenter, ASSURE Trial. Am. J. Cardiovasc. Drugs 2016, 16, 55–65. [Google Scholar] [CrossRef]
- Siedlecki, P.; Garcia, B.R.; Musch, T.; Brueckner, B.; Suhai, S.; Lyko, F.; Zielenkiewicz, P. Discovery of two novel, small-molecule inhibitors of DNA methylation. J. Med. Chem. 2006, 49, 678–683. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Tanaka, R.; Hamada, S.; Nakagawa, H.; Miyata, N. Design, synthesis, inhibitory activity, and binding mode study of novel DNA methyltransferase 1 inhibitors. Bioorganic Med. Chem. Lett. 2010, 20, 1124–1127. [Google Scholar] [CrossRef]
- Lee, B.H.; Yegnasubramanian, S.; Lin, X.; Nelson, W.G. Procainamide is a specific inhibitor of DNA methyltransferase 1. J. Biol. Chem. 2005, 280, 40749–40756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortiz, M.; Martín, A.; Arribas, F.; Coll-Vinent, B.; Del, A.C.; Peinado, R.; Almendral, J. Randomized comparison of intravenous procainamide vs. intravenous amiodarone for the acute treatment of tolerated wide QRS tachycardia: The PROCAMIO study. Eur. Heart J. 2017, 38, 1329–1335. [Google Scholar] [CrossRef] [Green Version]
- Newton, A.S.; Faver, J.C.; Micevic, G.; Muthusamy, V.; Kudalkar, S.N.; Bertoletti, N.; Anderson, K.S.; Bosenberg, M.W.; Jorgensen, W.L. Structure-Guided Identification of DNMT3B Inhibitors. ACS Med. Chem. Lett. 2020, 11, 971–976. [Google Scholar] [CrossRef]
- Hung, C.H.; Chan, S.H.; Chu, P.M.; Tsai, K.L. Quercetin is a potent anti-atherosclerotic compound by activation of SIRT1 signaling under oxLDL stimulation. Mol. Nutr. Food Res. 2015, 59, 1905–1917. [Google Scholar] [CrossRef]
- Dower, J.I.; Geleijnse, J.M.; Gijsbers, L.; Schalkwijk, C.; Kromhout, D.; Hollman, P.C. Supplementation of the Pure Flavonoids Epicatechin and Quercetin Affects Some Biomarkers of Endothelial Dysfunction and Inflammation in (Pre)Hypertensive Adults: A Randomized Double-Blind, Placebo-Controlled, Crossover Trial. J. Nutr. 2015, 145, 1459–1463. [Google Scholar] [CrossRef] [Green Version]
- Catalgol, B.; Batirel, S.; Taga, Y.; Ozer, N.K. Resveratrol: French paradox revisited. Front. Pharmacol. 2012, 3, 141. [Google Scholar] [CrossRef] [Green Version]
- Fukao, H.; Ijiri, Y.; Miura, M.; Hashimoto, M.; Yamashita, T.; Fukunaga, C.; Oiwa, K.; Kawai, Y.; Suwa, M.; Yamamoto, J. Effect of trans-resveratrol on the thrombogenicity and atherogenicity in apolipoprotein E-deficient and low-density lipoprotein receptor-deficient mice. Blood Coagul. Fibrinolysis 2004, 15, 441–446. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Wang, X.; Bi, S.; Pan, Z.; Liu, S.; Yu, H.; Lu, H.; Lin, X.; Wang, X.; Ma, T.; et al. Inhibitory effects of resveratrol on foam cell formation are mediated through monocyte chemotactic protein-1 and lipid metabolism-related proteins. Int. J. Mol. Med. 2014, 33, 1161–1168. [Google Scholar] [CrossRef] [Green Version]
- Beher, D.; Wu, J.; Cumine, S.; Kim, K.W.; Lu, S.C.; Atangan, L.; Wang, M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 2009, 74, 619–624. [Google Scholar] [CrossRef]
- Mansur, A.P.; Roggerio, A.; Goes, M.F.S.; Avakian, S.D.; Leal, D.P.; Maranhao, R.C.; Strunz, C.M.C. Serum concentrations and gene expression of sirtuin 1 in healthy and slightly overweight subjects after caloric restriction or resveratrol supplementation: A randomized trial. Int. J. Cardiol. 2017, 227, 788–794. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, B.; Campen, M.J.; Channell, M.M.; Wherry, S.J.; Varamini, B.; Davis, J.G.; Baur, J.A.; Smoliga, J.M. Resveratrol for primary prevention of atherosclerosis: Clinical trial evidence for improved gene expression in vascular endothelium. Int. J. Cardiol. 2013, 166, 246–248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chekalina, N.I. Resveratrol has a positive effect on parameters of central hemodynamics and myocardial ischemia in patients with stable coronary heart disease. Wiad. Lek. 2017, 70, 286–291. [Google Scholar] [PubMed]
- Lin, X.L.; Liu, M.H.; Hu, H.J.; Feng, H.R.; Fan, X.J.; Zou, W.W.; Pan, Y.Q.; Hu, X.M.; Wang, Z. Curcumin enhanced cholesterol efflux by upregulating ABCA1 expression through AMPK-SIRT1-LXRα signaling in THP-1 macrophage-derived foam cells. DNA Cell Biol. 2015, 34, 561–572. [Google Scholar] [CrossRef]
- Mohammadian, H.S.; Karimzadeh, M.R.; Azhdari, S.; Vahedi, P.; Abdollahi, E.; Momtazi-Borojeni, A.A. Modulatory effects of curcumin on the atherogenic activities of inflammatory monocytes: Evidence from in vitro and animal models of human atherosclerosis. Biofactors 2020, 46, 341–355. [Google Scholar] [CrossRef]
- Bhaskar, S.; Sudhakaran, P.R.; Helen, A. Quercetin attenuates atherosclerotic inflammation and adhesion molecule expression by modulating TLR-NF-kB signaling pathway. Cell. Immunol. 2016, 310, 131–140. [Google Scholar] [CrossRef]
- Stein, S.; Matter, C.M. Protective roles of SIRT1 in atherosclerosis. Cell Cycle 2011, 10, 640–647. [Google Scholar] [CrossRef] [Green Version]
- Peters, L.J.F.; Biessen, E.A.L.; Hohl, M.; Weber, C.; van der Vorst, E.P.C.; Santovito, D. Small Things Matter: Relevance of MicroRNAs in Cardiovascular Disease. Front. Physiol. 2020, 11, 793. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Gao, X.M.; Winbanks, C.E.; Boey, E.J.; Tham, Y.K.; Kiriazis, H.; Gregorevic, P.; Obad, S.; Kauppinen, S.; Du, X.J.; et al. Therapeutic inhibition of the miR-34 family attenuates pathological cardiac remodeling and improves heart function. Proc. Natl. Acad. Sci. USA 2012, 109, 17615–17620. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, C.; Sharma, A.R.; Sharma, G.; Lee, S.S. Therapeutic advances of miRNAs: A preclinical and clinical update. J. Adv. Res. 2021, 28, 127–138. [Google Scholar] [CrossRef]
- Abplanalp, W.T.; Fischer, A.; John, D.; Zeiher, A.M.; Gosgnach, W.; Darville, H.; Montgomery, R.; Pestano, L.; Allee, G.; Paty, I.; et al. Efficiency and Target Derepression of Anti-miR-92a: Results of a First in Human Study. Nucleic Acid Ther. 2020, 30, 335–345. [Google Scholar] [CrossRef]
- Nguyen, M.A.; Wyatt, H.; Susser, L.; Geoffrion, M.; Rasheed, A.; Duchez, A.C.; Cottee, M.L.; Afolayan, E.; Farah, E.; Kahiel, Z.; et al. Delivery of MicroRNAs by Chitosan Nanoparticles to Functionally Alter Macrophage Cholesterol Efflux in Vitro and in Vivo. ACS Nano 2019, 13, 6491–6505. [Google Scholar] [CrossRef] [PubMed]
- Skuratovskaia, D.; Vulf, M.; Komar, A.; Kirienkova, E.; Litvinova, L. Promising Directions in Atherosclerosis Treatment Based on Epigenetic Regulation Using MicroRNAs and Long Noncoding RNAs. Biomolecules 2019, 9, 226. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Yang, B.; Yang, H.; Wang, L.; Li, H.; Chen, S.; Lu, X.; Gu, D. MicroRNA-320b Modulates Cholesterol Efflux and Atherosclerosis. J. Atheroscler. Thromb. 2021. [Google Scholar] [CrossRef]
- Chen, Y.; Song, Y.; Huang, J.; Qu, M.; Zhang, Y.; Geng, J.; Zhang, Z.; Liu, J.; Yang, G.Y. Increased Circulating Exosomal miRNA-223 Is Associated with Acute Ischemic Stroke. Front. Neurol. 2017, 8, 57. [Google Scholar] [CrossRef] [Green Version]
- Devaux, Y.; Vausort, M.; Zhang, L.; Wagner, D.; Squire, I. Compositions and Methods for Evaluating Heart Failure. Patent EP2925884B1, 31 January 2018. [Google Scholar]
- Hong, Y.F.; Kim, H.; Kim, H.S.; Park, W.J.; Kim, J.Y.; Chung, D.K. Lactobacillus acidophilus K301 Inhibits Atherogenesis via Induction of 24 (S), 25-Epoxycholesterol-Mediated ABCA1 and ABCG1 Production and Cholesterol Efflux in Macrophages. PLoS ONE 2016, 11, e0154302. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Yang, Y.; Sun, X.; Tian, F.; Guo, S.; Wang, W.; Tian, Z.; Jin, H.; Zhang, Z.; Tian, Y. Sonodynamic therapy-induced foam cells apoptosis activates the phagocytic PPARγ-LXRα-ABCA1/ABCG1 pathway and promotes cholesterol efflux in advanced plaque. Theranostics 2018, 8, 4969–4984. [Google Scholar] [CrossRef] [PubMed]
- Javadifar, A.; Rastgoo, S.; Banach, M.; Jamialahmadi, T.; Johnston, T.P.; Sahebkar, A. Foam Cells as Therapeutic Targets in Atherosclerosis with a Focus on the Regulatory Roles of Non-Coding RNAs. Int. J. Mol. Sci. 2021, 22, 2529. [Google Scholar] [CrossRef]
- Shen, Z.; She, Q. Association between the Deletion Allele of Ins/Del Polymorphism (Rs145204276) in the Promoter Region of GAS5 with the Risk of Atherosclerosis. Cell. Physiol. Biochem. 2018, 49, 1431–1443. [Google Scholar] [CrossRef]
- Vausort, M.; Wagner, D.R.; Devaux, Y. Long noncoding RNAs in patients with acute myocardial infarction. Circ. Res. 2014, 115, 668–677. [Google Scholar] [CrossRef] [Green Version]
- Salgado-Somoza, A.; Zhang, L.; Vausort, M.; Devaux, Y. The circular RNA MICRA for risk stratification after myocardial infarction. Int. J. Cardiol. Heart Vasc. 2017, 17, 33–36. [Google Scholar] [CrossRef]
- Zhao, Z.; Li, X.; Gao, C.; Jian, D.; Hao, P.; Rao, L.; Li, M. Peripheral blood circular RNA hsa_circ_0124644 can be used as a diagnostic biomarker of coronary artery disease. Sci. Rep. 2017, 7, 39918. [Google Scholar] [CrossRef] [Green Version]
- Burd, C.E.; Jeck, W.R.; Liu, Y.; Sanoff, H.K.; Wang, Z.; Sharpless, N.E. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010, 6, e1001233. [Google Scholar] [CrossRef] [PubMed]
- Holdt, L.M.; Stahringer, A.; Sass, K.; Pichler, G.; Kulak, N.A.; Wilfert, W.; Kohlmaier, A.; Herbst, A.; Northoff, B.H.; Nicolaou, A.; et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 2016, 7, 12429. [Google Scholar] [CrossRef] [Green Version]
- Vausort, M.; Salgado-Somoza, A.; Zhang, L.; Leszek, P.; Scholz, M.; Teren, A.; Burkhardt, R.; Thiery, J.; Wagner, D.R.; Devaux, Y. Myocardial Infarction-Associated Circular RNA Predicting Left Ventricular Dysfunction. J. Am. Coll. Cardiol. 2016, 68, 1247–1248. [Google Scholar] [CrossRef]

| Expression Regulation Levels (Cellular Compartments) | Participants | ABCA1 | ABCG1 | SCARB1 |
|---|---|---|---|---|
| Genome (nucleus) | ||||
| SNPs/mutations | ● | ● | ● | |
| Transcription (nucleus) | ||||
| methylation of the promoter region | ● | ● | ● | |
| transcription activators | ● | ● | ● | |
| transcription repressors | ● | ● | ||
| transcription factors | ● | ● | ● | |
| lncRNAs | ● | ● | ● | |
| Post-transcriptional regulation (cytoplasm) | lncRNAs interacting with miRNA | ● | ||
| lncRNAs interacting with proteins or DNA | ● | ● | ||
| miRNAs | ● | ● | ● | |
| circRNA | ● |
| miRNA | Target | Expression Change in Cardiovascular Diseases (CVD) and Knockout and Model Mice | In Vitro Effect on Lipid Level and Reverse Cholesterol Transport (RCT) | In Vivo Effect on Lipid Level, RCT and Atherosclerosis |
|---|---|---|---|---|
| miR-9 | ABCA1 | Plasma level of hsa-miR-9-3p decreased in patients with unstable angina (UA) [99]. | MiR-9-5p directly bound to the 3′-UTR of ABCA1 and reduced its mRNA and protein levels in macrophages [100]. | |
| miR-10b | ABCA1/ABCG1 | MiR-10b level increased in atherosclerotic plaques in humans [101]. | MiR-10b directly bound to the 3′-UTR of ABCA1/ABCG1 and suppressed their expression and cholesterol efflux from mouse peritoneal macrophages (MPMs) and human THP-1 monocytes [102]. | In ApoE−/− mice, miR-10b suppressed the expression of ABCA1/ABCG1 and RCT from macrophages to feces, thus contributing to the development of atherosclerosis, the growth of plaques and their instability in the late stages [102,103]. |
| miR-17 | ABCA1 | An increase in the level of miR-17-5p has been found in leukocytes of patients with atherosclerosis [104], in plasma of patients with UA [105], acute myocardial infarction (AMI) [106], CAD [107,108]. The serum level of miR-17-5p was also associated with the development of ischemic heart disease (IHD) [109] and the severity of CAD [110]. miR-17-3p levels also increased in atherosclerotic plaques in humans [101]. However, a decrease in the circulating miR-17-5p level has been found in patients with CAD [111] and CHD [112]. | MiR-17-5p directly bound to the 3′-UTR of ABCA1 and suppressed its expression in mouse macrophage RAW264.7 [104]. | The level of miR-17-5p increased in the macrophages of ApoE−/− mice on a high-cholesterol diet [104]. |
| miR-19b | ABCA1 | MiR-19b levels elevated in human atherosclerotic plaques and rat aortic tissues of the abdominal aortic aneurysm (AAA) model [113,114], in plasma of patients with AMI [115] and in plasma endothelial microparticles (EMPs) of patients with UA [116]. | MiR-19b directly suppressed ABCA1 expression and cholesterol efflux from MPMs and macrophages derived from human THP-1 monocytes [117]. | In ApoE−/− mice, miR-19b suppressed the expression of ABCA1, RCT and the level of HDL in plasma, thus increasing the size of aortic plaques and contributing to the development of atherosclerosis [117,118]. |
| miR-20a/b | ABCA1 | Changes in miR-20a expression in atherosclerosis-associated diseases are multidirectional. Thus, the level of miR-20a increased in human aorta with AAA [119] and in plasma of patients with UA as well [99,105]. In contrast, the level of miR-20a decreased in blood cells of patients with AMI [120] and in plasma of patients with CAD [111]. MiR-20b was also low in blood cells of patients with the peripheral arterial disease (PAD) [121]. Expression of miR-20a/b decreased in the liver of ApoE−/− mice on a high fat diet [98]. | MiR-20a/b bound to the 3′-UTR of ABCA1 and suppressed its expression and cholesterol efflux from THP-1- and RAW 264.7-derived foam cells [98]. | In ApoE−/− mice, miR-20a/b reduced ABCA1 expression in the liver, RCT efficiency and HDL synthesis, thus contributing to the development of atherosclerosis [98]. |
| miR-23a | ABCA1ABCG1 | Increased values for miR-23a were associated with atherosclerosis-related diseases, i.e., an increased miR-23a level has been detected in the plasma of patients with acute ischemic stroke (AIS) with vulnerable carotid plaques [122], in plasma of patients with UA [99] and in plasma and PBMCs of patients with CAD [123,124,125,126]. miR-23a levels are correlated with plaque development [122], stenosis degree [123] and poor clinical outcomes in CAD [124]. OxLDL upregulated miR-23a expression in macrophages [122]. However, miR-23a level in plasma decreased within 24 h of stroke onset in humans [127]. | MiR-23a suppressed the activity of 3′-UTR of ABCA1 and ABCG1, reduced their expression and cholesterol efflux, that led to foam cell formation [122]. | In ApoE−/− mice, miR-23a suppressed ABCA1 and ABCG1 expression, promoted atherosclerosis and increased plaque vulnerability [122]. |
| miR-24 | SCARB1 | The data are contradictory. Fatty acids increased the expression of miR-24 in HepG2 cells. The miR-24 levels significantly increased in the liver of obese mice [128], in the plasma of patients with stable angina pectoris (AP) [129], in PBMCs of patients with CAD [130]. However, miR-24 levels reduced in blood of patients with atherosclerosis [131] and in plasma of patients with familial hypercholesterolemia (FH) [132]. | MiR-24 directly suppressed the expression of SR-BI by binding to the 3′-UTR of mRNA, thus reducing the selective uptake of HDL-CE by HepG2 and THP-1 cells [128,133]. In addition, steroidogenesis reduced in steroidogenic cells [128]. | In ApoE−/− mice, miR-24 reduced the expression of SR-BI and promoted the formation of atherosclerotic plaques [133]. |
| miR-26a/b | ABCA1 | The level of miR-26a-1 increased in plasma of patients with AMI [134]. The level of miR-26b increased in plasma of patients with UA [99], while miR-26a/b increased in EMPs of patients with UA [116]. Moreover, the expression of miR-26b was significantly upregulated in atherosclerotic plaques in humans [101]. However, miR-26b decreased in blood cells of patients with peripheral arterial disease (PAD) [121]. | In RAW 264.7, THP-1, HEK293T and HepG2 cells, miR-26 bound to the 3′-UTR of ABCA1 and suppressed its expression [135]. | |
| miR-27a/b | ABCA1 | The level of miR-27a increased in PBMCs of patients with CAD [32] and in plasma of patients with UA [99]. The level of miR-27b significantly increased in sclerotic intima samples and in serum of patients with atherosclerosis obliterans [136], in plasma of patients with AAA [137], as well as in PBMCs of the patients with CAD, and expression levels of miR-27b were significantly correlated with the severity of stenosis [123]. The level of miR-27b elevated in the liver of C57BL/6J mice, as well as in ApoE−/− female mice on a high-fat “Western” diet [138]. However, the decreased levels of miR-27b were observed in blood cells of patients with PAD [121] and in plasma of patients with CAD [111], as well as in aneurysm tissues of patients with AAA [137]. A reduced level of miR-27b is associated with heart failure, atherosclerosis, and the severity of PAD symptoms [139]. | MiR-27a/b directly targeted the 3′-UTR of ABCA1, significantly reducing its mRNA and protein levels in foam cells derived from THP-1 and RAW 264.7, as well as in HepG2 cells [140]. MiR-27a/b also reduced cholesterol efflux from THP-1 macrophages to apoA-I through the suppression of ABCA1. A similar effect of miR-27b on ABCA1 mRNA and protein levels and cholesterol efflux existed for Huh7 cells [141]. | Modulation of miR-27b expression in wild-type mice regulated ABCA1 expression in the liver but does not affect lipid levels [141]. |
| miR-28 | ABCA11 | The level of miR-28-5p increased in patients with UA [142,143]. | miR-28-5p targeted the signal-regulated kinase 2 (ERK2) and inhibited its expression that led to increase of ABCA1 expression in THP-1 derived macrophages and HepG2 cells [142,143]. | |
| miR-30e | ABCA1 | The expression of miR-30e was significantly upregulated in the serum exosome of patients with CAD [26], in atherosclerotic plaques in humans [101], in plasma of patients with UA [99], and in blood cells of patients with AMI [120]. Moreover, miR-30e is considered as a differential biomarker for AMI [144]. However, there is evidence that miR-30e expression reduced in PBMCs of patients with lower extremities arterial disease (LEAD) [145] and in the whole blood of CAD patients [146]. | MiR-30e directly targeted 3′-UTR of ABCA1 and suppressed its protein expression [26]. | |
| miR-34a | ABCA1/ABCG1 | All studies evidence the increase of miR-34a in atherosclerosis- associated diseases. Thus, the level of miR-34a significantly increased in atherosclerotic plaques in humans and in ApoE−/− mice [147,148], in PBMCs of patients with LEAD [145], in plasma of patients with CAD [126,149] and AP [129]. Upregulated miR-34a is considered as a universal marker for AMI and UA [144]. | In HepG2 cells, miR-34a directly interacted with the 3′-UTR of ABCA1 and ABCG1 mRNA and suppressed their expression [147]. Moreover, miR-34a inhibited cholesterol efflux from THP-1 and MPMs cells. | In mice, the downregulation of ABCA1 and ABCG1 by miR-34a promoted RCT suppression to plasma, liver and feces [147]. In ApoE−/− and Ldlr−/− mice, miR-34a promoted dyslipidemia, plaque growth, and instability. |
| miR-92a | ABCA1 | The data on miR-92a expression in atherosclerosis are contradictory. The increased level of miR-92a was found in plasma and plasma exosomes of patients with the initial stage of atherosclerosis [150], with CAD [26,151], in aneurysm tissues of AAA [119], in human coronary atherosclerotic plaques [114], in plasma of patients with hypertension, especially with thickening of the carotid artery wall [152], in plasma of patients with UA [105], and with asymptomatic carotid artery stenosis, where it was correlated with the degree of stenosis [153], in PBMCs of CAD patients and in EMPs of patients with UA [116]. Moreover, upregulated miR-92a is considered as a differential biomarker for UA [144]. However, miR-92a expression decreased in the blood of patients with CAD [108,111,154], CHD [112] and atherosclerosis [155], in plasma and atherosclerotic plaques in PAD patients with cardiovascular events (CVEs) [156]. | miR-92a directly targeted 3′-UTR of ABCA1 and suppressed its protein expression [26]. | Increased expression of miR-92a contributed to the development of atherosclerotic plaques under the influence of oxLDL in Ldlr−/− mice [157]. |
| miR-93 | ABCA1 | Mostly, miR-93 levels increased in atherosclerosis. Thus, increased miR-93-5p level was detected in plasma of patients with critical coronary stenosis [158], with UA [105], CAD [159] and in blood cells of patients with AMI [120]. Moreover, miR-93 is considered as a universal biomarker for both AMI and UA [144]. However, miR-93 level decreased in CAD patients [160]. | miR-93 directly targeted 3′-UTR of ABCA1 and suppressed its protein expression [160]. | |
| miR-96 | SCARB1 | MiR-96 level decreased in ApoE−/− mice on a high-fat diet [161]. The level of miR-96 was significantly upregulated in THP-1 cells stimulated to differentiate into macrophages. | miR-96 directly targeted 3′-UTR of SCARB1, suppressed its protein expression and HDL-C uptake by HepG2 and other human liver cells [161]. However, miR-96 increased HDL-C uptake by THP-1 cells, probably through the regulation of other pathways of cholesterol delivery. | |
| miR-101 | ABCA1 | IL-6 and TNF-α induced miR-101 expression in HepG2 cells and THP-1 macrophages [162]. During inflammation, miR-101 may promote the intracellular accumulation of lipids, which results in atherosclerosis. | MiR-101 directly interacted with the 3′-UTR of ABCA1 and suppressed its protein expression, that reduced cholesterol efflux from cells to apoA-I [162]. | |
| miR-106b | ABCA1 | Level of miR-106b significantly decreased in plasma of patients with CAD and was correlated with HDL level [108]. MiR-106b level increased in plasma microparticles (MPs) of UA patients [105]. | MiR-106b directly bound to the 3′-UTR of ABCA1 and repressed its translation [163]. In neuronal cells (Neuro2a), miR-106b reduced ABCA1 levels and cholesterol efflux. | |
| miR-125a | SCARB1 | miR-125a level decreased in the coronary arteries of patients with atherosclerotic plaques [164] and in the serum of patients with atherosclerosis [165] but increased in atherosclerotic plaques [101]. | MiR-125a directly targeted 3′-UTR of SCARB1 and suppressed SR-BI expression [166]. In rat/mouse Leydig tumor cells, suppression of SR-BI expression at mRNA and protein levels under the influence of miR-125a led to a decrease of HDL-CE uptake by cells and a decrease in HDL-dependent progesterone production. In mouse Hepa1-6 cells, miR-125a also suppressed SR-BI expression and HDL-CE uptake. However, in HepG2 cells, such effect of miR-125a was not found [161]. | |
| miR-128 | ABCA1/ABCG1 | In mice on a high-fat diet, the level of miR-128 decreased in the liver, brain, and kidneys [167] but increased in the blood, brain, and heart [168]. miR-128-2 may prevent cholesterol efflux from cells at low cholesterol [167]. | MiR-128-2 targeted 3′-UTR of ABCA1 and ABCG1 and inhibited their expression that led to the suppression of cholesterol efflux from HepG2, MCF7, and HEK293T cells [167]. Similar effects for miR-128-1 were found in mouse macrophages [169]. | miR-128 is inversely correlated with ABCA1 and ABCG1 expression levels in different tissues of mice on a high-fat diet [167]. |
| miR-130b | ABCA1 | MiR-130b directly interacted with the 3′-UTR of ABCA1 and suppressed its expression in HepG2 and in mouse macrophages, that led to reducing the cholesterol efflux [169]. | ||
| miR-143 | ABCA1 | MiR-143 was up-regulated and ABCA1 was down-regulated in PAH patients [170]. MiR-143 level increased in human coronary atherosclerotic plaques [114]. | MiR-143 directly suppressed the expression of ABCA1 in pulmonary artery smooth muscle cells (PASMCs) [170]. | MiR-143 promoted the development of hypoxia-induced pulmonary arterial hypertension (PAH) in vivo, presumably due to its influence on ABCA1 expression [170]. The studies with Ldlr−/− and Ldlr−/− miR-143/145−/− double knockout mice revealed the contribution of these miRNAs to the development of atherosclerosis [171]. |
| miR-144 | ABCA1 | MiR-144 increased in the plasma of patients with UA [99] and CAD [149,172,173], in monocytes of patients with hypertension [174]. However, miR-144 level was decreased in AAA tissue [137]. The level of miR-144 was associated with AMI [175]. LXR ligands increased the expression of miR-144 in mouse and human liver cells and macrophages, that may be important in homeostasis [176]. FXR transactivated miR-144 which suppressed ABCA1 and cholesterol efflux [177]. | MiR-144 directly interacted with the 3′-UTR of ABCA1 and decreased its expression and cholesterol efflux to apoA-I [175,176,178]. | miR-144 reduced the levels of ABCA1 and HDL in the liver and plasma of mice [176,177]. In ApoE−/− mice, miR-144-3p decreased plasma HDL levels, impaired RCT and promoted the development of atherosclerosis [175]. A high-fat diet induced the development of atherosclerosis in miR-144−/− mice [179]. miR-144 promoted lipid accumulation and lipid disorder in F1-zebrafish [180]. |
| miR-145 | ABCA1 | Data are contradictory. The level of miR-145 increased in the blood of patients with PAH [170], in plasma of patients with AMI [106] and within 24 h of stroke onset [127]. Upregulated level of miR-145 is considered as a biomarker for both AMI and UA [144]. The miR-145 levels are correlated with the size of the infarction area and may predict a long-term clinical outcome after AMI [181]. However, level of miR-145 decreased in the plasma of patients with AMI [182] and in the plasma and blood of patients with CAD, including very early onset [183], where it is correlated with disease severity [111,146,184]. | MiR-145 targeted 3′-UTR of ABCA1 and suppressed its protein expression and cholesterol efflux from HepG2 cells [178]. | MiR-145 promoted a decrease in the ABCA1 protein in the mouse pancreas, as well as an increase in total cholesterol levels and a decrease in insulin secretion [178]. The studies in Ldlr−/− and Ldlr−/− miR-143/145−/− double knockout mice showed the contribution of these miRNAs to the development of atherosclerosis [171]. |
| miR-148 | ABCA1 | The expression of miR-148b reduced in the serum of patients with atherosclerosis and in human aortic smooth muscle cells stimulated by ox-LDL [185]. The level of miR-148-3p increased in the liver of rhesus monkeys on a high-fat diet, as well as in mice (ob/ob) with genetically determined obesity [186]. | MiR-148 directly bound the 3′-UTR of ABCA1 and suppressed its expression [169,178,186]. As a result, miR-148 suppressed cholesterol efflux from HepG2 and mouse macrophages [169]. | In C57BL/6J and ApoE−/− mice on a high-fat diet, miR-148 reduced liver ABCA1 and blood HDL [169]. In Ldlr−/− mice on a high-fat diet, miR-148 contributed to a decrease of ABCA1 in the liver and HDL in blood [186]. |
| miR-183 | ABCA1 | In macrophages derived from THP-1, IL-18 promoted an increase in miR-183 expression with a concomitant decrease in ABCA1 expression and cholesterol efflux, which may contribute to the development of atherosclerosis [187]. | MiR-183 directly interacted with the 3′-UTR of ABCA1 and suppressed its expression [187]. | |
| miR-185 | SCARB1 | MiR-185-3p was upregulated in atherosclerotic mouse aorta [188]. miR-185 also increased in atherosclerotic plaques in humans [101]. However, in the liver of ApoE−/− mice on a high-fat diet, the miR-185 level decreased [161]. | MiR-185 directly interacted with the 3′-UTR of SCARB1 and suppressed the expression of SR-BI and HDL-C uptake in THP-1 cells and human hepatic cell lines [161]. | |
| miR-188 | ABCA1 | MiR-188-3p decreased in ApoE−/− mice with atherosclerosis [189]. | In ApoE−/− mice with atherosclerosis, miR-188-3p upregulated ABCA1 level in serum and promoted a decrease of lipid accumulation within the vessels and atherosclerosis [189]. | |
| miR-212 | ABCA11 | The miR-212 level decreased in plaques and macrophages of ApoE−/− mice on a high-fat diet [190]. | In THP-1 macrophages, miR-212 targeted SIRT1, which led to inhibition of ABCA1 expression, decreased cholesterol efflux and increased intracellular lipid accumulation [190]. | |
| miR-223 | SCARB1/ABCA11 | miR-223 increased in CVD i.e., in ApoE−/− mice [191], in serum, in the vascular wall of patients with atherosclerosis obliterans [192], in the plasma of patients with AMI [115], PAD with cardiovascular events (CVEs) [156], unstable coronary artery disease (UCAD) [193], coronary artery calcification (CAC) [194] and UA [99,105], in platelets of patients with CAD [195], in atherosclerotic plaques of patients with PAD with cardiovascular events (CVEs) [156], and in aneurysm tissues of patients with AAA [119]. HDL-transported miR-223 elevated in patients with hypercholesterolemia and in Ldlr−/− and ApoE−/− mice on a high-fat diet. miR-223 increased in human hepatocytes with a high level of extracellular cholesterol [196]. An increased miR-223 level is associated with an increased risk of CVD [196]. MiR-223 expression is associated with atherogenesis in CAD [197]. However, the expression of miR-223 decreased in PBMCs of patients with CAD with the lowest stenosis less than 50% [198]. A reduced level of miR-223 is associated with heart failure, atherosclerosis, and the severity of PAD symptoms [139]. In THP-1 macrophages, miR-223 expression was significantly upregulated bur had no effect on SCARB1 and HDL-C uptake [161]. A reduced cholesterol level caused a decrease in the level of miR-223 in J774 macrophages and Huh7 cells [199]. | MiR-223 directly targeted the 3′-UTR of SCARB1, suppressed SR-B1 expression and the uptake of HDL-C in human hepatic cells [161,199]. miR-223 targeted Sp3, the repressor of Sp1-directed ABCA1 transcription. Thus, miR-223 promoted the indirect increase of mRNA and protein levels of ABCA1, as well as the cholesterol efflux to apoA-I in Huh7 cells [199]. | In miR-223−/− mice the level of SR-BI in the liver reduced, but total cholesterol and HDL-C increased in plasma. Cholesterol level increased in the liver of these mice [199]. |
| miR-301b | ABCA1 | MiR-301b directly bound to the 3′-UTR of ABCA1 and suppressed its expression in HepG2 and mouse macrophages, that led to a decrease of cholesterol efflux [169]. | ||
| miR-302a | ABCA1 | Ox-LDL downregulated miR-302a expression in mouse macrophages [200]. In the liver of Ldlr−/− mice on Western-type diet, miR-302a decreased [201]. | MiR-302a targeted 3′-UTR of ABCA1 and suppressed its protein expression in primary mouse and human macrophages, leading to suppression of cholesterol efflux [200]. | In Ldlr−/− mice on an atherogenic diet, miR-302a suppressed ABCA1 expression in the liver and aorta with a decrease of plasma HDL level, that promoted the growth of plaques, their instability and inflammation [200]. |
| miR-361-5p | ABCA1 | MiR-361-5p directly bound to the 3′-UTR of ABCA1 and suppressed its expression [202]. | ||
| miR-378 | ABCG1 | MiR-378 levels increased in aortas during the progression of atherosclerosis in ApoE−/−mice [203]. Plasma miR-378 expression was significantly downregulated in patients with CAD [146,204], CHD [112]. Moreover, it is considered as biomarker for risk and severity of CHD [112]. | MiR-378 directly interacted with the 3′-UTR of ABCG1 and suppressed its expression that led to downregulation of cholesterol efflux from mouse and human macrophages [203]. | In ApoE−/− mice, miR-378 presumably downregulated ABCG1 expression in peritoneal macrophages, leading to decreased RCT and atherosclerosis progression [203]. |
| miR-486 | ABCA11 | The level of miR-486 increased in the plasma of obese children and is associated with body mass index and other indicators of obesity [205]. The level of miR-486 elevated in the blood of patients with CAD [151] and is associated with the risk of developing cardiovascular diseases [109,206]. | MiR-486 directly bound to 3′-UTR of histone acetyltransferase-1 (HAT1) and suppressed its expression with a concomitant decrease in ABCA1 expression at both mRNA and protein level, that led to cholesterol accumulation in THP-1 cells [207]. | |
| miR-613 | ABCA1 | PPAR-γ, which induces the expression of a cascade of genes involved in cholesterol efflux from macrophages, negatively regulated the expression of miR-613 at transcriptional level [208]. | miR-613 targeted 3′-UTR of ABCA1 and suppressed its protein expression, which led to inhibition of cholesterol efflux from THP-1 cells activated by PPAR-γ [208]. | |
| miR-758 | ABCA1 | The level of miR-758 decreased in cholesterol-enriched macrophages, as well as in pancreatic macrophages and liver cells in mice on a high-fat diet [209]. The level of miR-758 increased in plaques from patients with hypercholesterolemia compared to plaques of patients with normal cholesterol [210]. | MiR-758 directly interacted with 3′-UTR of ABCA1, suppressed its expression and cholesterol efflux to apoA-I in mouse and human macrophages [209] and HepG2 cells [211]. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rozhkova, A.V.; Dmitrieva, V.G.; Nosova, E.V.; Dergunov, A.D.; Limborska, S.A.; Dergunova, L.V. Genomic Variants and Multilevel Regulation of ABCA1, ABCG1, and SCARB1 Expression in Atherogenesis. J. Cardiovasc. Dev. Dis. 2021, 8, 170. https://doi.org/10.3390/jcdd8120170
Rozhkova AV, Dmitrieva VG, Nosova EV, Dergunov AD, Limborska SA, Dergunova LV. Genomic Variants and Multilevel Regulation of ABCA1, ABCG1, and SCARB1 Expression in Atherogenesis. Journal of Cardiovascular Development and Disease. 2021; 8(12):170. https://doi.org/10.3390/jcdd8120170
Chicago/Turabian StyleRozhkova, Alexandra V., Veronika G. Dmitrieva, Elena V. Nosova, Alexander D. Dergunov, Svetlana A. Limborska, and Liudmila V. Dergunova. 2021. "Genomic Variants and Multilevel Regulation of ABCA1, ABCG1, and SCARB1 Expression in Atherogenesis" Journal of Cardiovascular Development and Disease 8, no. 12: 170. https://doi.org/10.3390/jcdd8120170
APA StyleRozhkova, A. V., Dmitrieva, V. G., Nosova, E. V., Dergunov, A. D., Limborska, S. A., & Dergunova, L. V. (2021). Genomic Variants and Multilevel Regulation of ABCA1, ABCG1, and SCARB1 Expression in Atherogenesis. Journal of Cardiovascular Development and Disease, 8(12), 170. https://doi.org/10.3390/jcdd8120170