PdMFS1 Transporter Contributes to Penicilliun digitatum Fungicide Resistance and Fungal Virulence during Citrus Fruit Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms and Culture Conditions
2.2. Nucleic Acids Isolation
2.3. Identification of PdMFS1
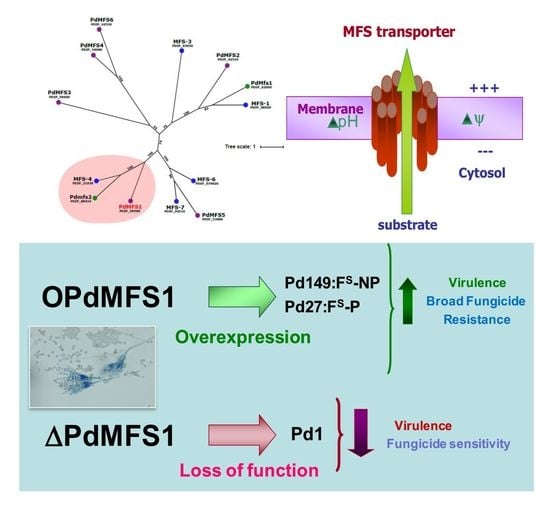
2.4. PdMFS1 Gene Deletion and Gene Overexpression
2.5. Virulence Tests
2.6. Fungicide Sensitivity Test
2.7. Quantitative Real-Time PCR
2.8. Statistical Analysis
3. Results
3.1. Analysis of PdMFS1
3.2. Generation of the PdMFS1 Mutants
3.3. Overexpression of PdMFS1 in Different P. digitatum Strains
3.4. Fungicide Sensitivity
3.5. Virulence Evaluation
3.6. Expression Pattern of PdMFS1
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Palou, L. Penicillium digitatum and Pencillium italicum (Green Mold, Blue Mold). In Postharvest Decay: Control Strategies; Bautista-Baños, S., Ed.; Elsevier: Amsterdam, The Netherlands, 2014; pp. 45–102. [Google Scholar]
- Kanetis, L.; Forster, H.; Adaskaveg, J.E. Baseline sensitivities for the new postharvest fungicides against Penicillium species on citrus and multiple resistance evaluations in P. digitatum. Plant Dis. 2008, 92, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Michailides, T.J. Advances in understanding molecular mechanisms of fungicide resistance and molecular detection of resistant genotypes in phytopathogenic fungi. Crop Prot. 2005, 24, 853–863. [Google Scholar] [CrossRef]
- Sánchez-Torres, P.; Tuset, J.J. Molecular insights into fungicide resistance in sensitive and resistant Penicillium digitatum strains infecting citrus. Postharvest Biol. Technol. 2011, 59, 159–165. [Google Scholar] [CrossRef]
- Sang, H.; Hulvey, J.P.; Green, R.; Xu, H.; Im, J.; Chang, T.; Jung, G. A xenobiotic detoxification pathway through transcriptional regulation in filamentous fungi. mBio 2018, 9, e00457-18. [Google Scholar] [CrossRef] [PubMed]
- Morschhauser, J. Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 2010, 47, 94–106. [Google Scholar] [CrossRef]
- Yan, N. Structural advances for the major facilitator superfamily (MFS) transporters. Trend Biochem. Sci. 2013, 38, 151–159. [Google Scholar] [CrossRef]
- Dos Santos, S.C.; Teixeira, M.C.; Dias, P.J.; Sá-Correia, I. MFS transporter required for multidrug/multixenobiotic (MD/MX) resistance in the model yeast: understanding their physiological function through postgenomic approaches. Front. Physiol. 2014, 5, 180. [Google Scholar] [CrossRef]
- Hayashi, K.; Schoonbek, H.J.; de Waard, M.A. Bcmfs1. A novel major facilitator superfamily tranporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol. 2002, 68, 4996–5004. [Google Scholar] [CrossRef]
- Choquer, M.; Lee, M.; Bau, H.; Chung, K. Deletion of a MFS transporter like gene in Cercospora nicotianae reduces cercosporin toxin accumulation and fungal virulence. FEMS Lett. 2007, 581, 489–494. [Google Scholar]
- Kretschmer, M.; Leroch, M.; Mosbach, A.; Walker, A.-S.; Fillinger, S.; Mernke, D.; Schoonbeek, H.-J.; Pradier, J.-M.; Leroux, P.; De Waard, M.A.; et al. Fungicide-Driven Evolution and Molecular Basis of Multidrug Resistance in Field Populations of the Grey Mould Fungus Botrytis cinerea. PLoS Pathog. 2009, 512, e1000696. [Google Scholar]
- Roohparvar, R.; De Waard, M.A.; Kema, G.H.; Zwiers, L.H. MgMfs1, a major facilitator superfamily transporter from the fungal wheat pathogen Mycosphaerella graminicola, is a strong protectant against natural toxic compounds and fungicides. Fungal Genet. Biol. 2007, 44, 378–388. [Google Scholar] [CrossRef] [PubMed]
- Roohparvar, R.; Mehrabi, R.; Van Nistelrooy, J.G.M.; Zwiers, L.; De Waard, M.A. The drug transporter MgMfs1 can modulate sensitivity of field strains of the fungal wheat pathogen Mycosphaerella graminicola to the strobilurin fungicide trifloxystrobin. Pest Manag. Sci. 2008, 64, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Omrane, S.; Sghyer, H.; Audéon, C.; Lanen, C.; Duplaix, C.; Walker, A.-S.; Fillinger, S. Fungicide efflux and the MgMFS1 transporter contribute to the multidrug resistance phenotype in Zymoseptoria tritici field isolates. Environ. Microbiol. 2015, 178, 2805–2823. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.H.; Tsai, H.C.; Chung, K.R. A Major Facilitator Superfamily Transporter-Mediated Resistance to Oxidative Stress and Fungicides Requires Yap1, Skn7, and MAP Kinases in the Citrus Fungal Pathogen Alternaria alternata. PLoS ONE 2017, 121, e0169103. [Google Scholar] [CrossRef]
- Wang, J.Y.; Sun, X.P.; Lin, L.Y.; Zhang, T.Y.; Ma, Z.H.; Li, H.Y. PdMfs1, a major facilitator superfamily transporter from Penicilliun digitatum, is partially involved in the imazalil-resistance and pathogenicity. African J. Microbiol. Res. 2012, 6, 95–105. [Google Scholar]
- Wu, Z.; Wang, S.; Yuan, Y.; Zhang, T.; Liu, J.; Liu, D. A novel major facilitator superfamily transporter in Penicillium digitatum (PdMFS2) is required for prochloraz resistance, conidiation and full virulence. Biotechnol. Lett. 2016, 38, 1349–1357. [Google Scholar] [CrossRef]
- De Ramón-Carbonell, M.; Sánchez-Torres, P. The transcription factor PdSte12 contributes to Penicillium digitatum virulence during citrus fruit infection. Postharvest Biol Technol. 2017, 125, 129–139. [Google Scholar] [CrossRef]
- Marcet-Houben, M.; Ballester, A.R.; de la Fuente, B.; Harries, E.; Marcos, J.F.; González-Candelas, L.; Gabaldón, T. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics 2012, 13, 646. [Google Scholar] [CrossRef]
- Prober, J.M.; Trainor, G.L.; Dam, R.J.; Hobbs, F.W.; Robertson, C.W.; Zagursky, R.J.; Cocuzza, A.J.; Jensen, M.A.; Baumeister, K. A system for rapid DNA sequencing with fluorescent chain terminating dideoxynucleotides. Science 1987, 238, 336–341. [Google Scholar] [CrossRef]
- López-Pérez, M.; Ballester, A.R.; González-Candelas, L. Identification and functional analysis of Penicillium digitatum genes putatively involved in virulence towards citrus fruit. Mol. Plant Pathol. 2015, 16, 262–275. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frandsen, R.J.N.; Andersson, J.A.; Kristensen, M.B.; Giese, H. Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol. Biol. 2008, 9, 70. [Google Scholar] [CrossRef] [PubMed]
- De Ramón-Carbonell, M.; Sánchez-Torres, P. Involvement of Penicillium digitatum PdSUT1 in fungicide sensitivity and virulence during citrus fruit infection. Microbiol. Res. 2017, 203, 57–67. [Google Scholar] [CrossRef] [PubMed]
- González-Candelas, L.; Alamar, S.; Sánchez-Torres, P.; Zacarías, L.; Marcos, J.F. A transcriptomic approach highlights induction of secondary metabolism in citrus fruit in response to Penicillium digitatum infection. BMC Plant Biol. 2010, 10, 194. [Google Scholar] [CrossRef] [PubMed]
- Ballester, A.R.; Marcet-Houben, M.; Levin, E.; Sela, N.; Selma-Lazaro, C.; Carmona, L.; Wisniewski, M.; Droby, S.; González-Candelas, L.; Gabaldón, T. Genome, transcriptome, and functional analyses of Penicillium expansum provide new insights into secondary metabolism and pathogenicity. Mol. Plant-Microbe Interact. 2015, 28, 232–248. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Wang, S.; Qin, T.; Li, N.; Niu, Y.; Li, D.; Yuan, Y.; Geng, H.; Xiong, L.; Liu, D. Whole transcriptome analysis of Penicillium digitatum strains treatmented with prochloraz reveals their drug-resistant mechanisms. BMC Genomics 2015, 16, 855. [Google Scholar] [CrossRef]
- Fluman, N.; Ryan, C.; Whitelegge, J.; Bibi, E. Dissection of mechanistic principles of a secondary multidrug efflux protein. Mol. Cell 2012, 47, 777–787. [Google Scholar] [CrossRef]
- Liu, M.; Liu, J.; Wang, W.M. Isolation and functional analysis of Thmfs1, the first major facilitator superfamily transporter from the biocontrol fungus Trichoderma harzianum. Biotechnol Lett. 2012, 34, 1857–1862. [Google Scholar] [CrossRef]
- Hayashi, K.; Schoonbeek, H.; Sugiura, H.; De Waard, M.A. Multidrug resistance in Botrytis cinerea associated with decreased accumulation of the azole fungicide oxpoconazole and increased transcription of the ABC transporter gene BcatrD. Pestic. Biochem. Physiol. 2001, 70, 168–179. [Google Scholar] [CrossRef]
- De Waard, M.A.; Andrade, A.C.; Hayashi, K.; Schoonbeek, H.J.; Stergiopoulos, I.; Zwiers, L.H. Impact of fungal drug transporters on fungicide sensitivity, multidrug resistance and virulence. Pest Manag. Sci. 2006, 62, 195–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamamoto, H.; Nawata, O.; Hasegawa, K.; Nakaune, R.; Lee, Y.J.; Makizumi, Y.; Akutsu, K.; Hibi, T. The role of the ABC transporter gene PMR1 in demethylation inhibitor resistance in Penicillium digitatum. Pestic. Biochem. Physiol. 2001, 70, 19–26. [Google Scholar] [CrossRef]
- Ghosoph, J.M.; Schmidt, L.S.; Margosan, D.A.; Smilanick, J.L. Imazalil resistance linked to a unique insertion sequence in the PdCYP51 promoter region of Penicillium digitatum. Postharvest Biol. Technol. 2007, 44, 9–18. [Google Scholar] [CrossRef]
- Hamamoto, H.; Hasegawa, K.; Nakaune, R.; Lee, Y.J.; Makizumi, Y.; Akutsu, K.; Hibi, T. Tandem repeat of a transcription enhancer upstream of the sterol 14_demethylase gene (CYP51) Penicillium digitatum. Appl. Environ. Microbiol. 2000, 66, 3421–3426. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, J.; Feng, D.; Ma, Z.; Li, H. PdCYP51B a new putative sterol 14 alpha demethylase gene of Penicillium digitatum involved in resistance to imazalil and other fungicides inhibiting ergosterol synthesis. Appl. Microbiol. Biotechnol. 2011, 91, 1107–1119. [Google Scholar] [CrossRef] [PubMed]
- Callahan, T.M.; Rose, M.S.; Meade, M.J.; Ehrenshaft, M.; Upchurch, R.G. CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol. Plant Microbe Interact. 1999, 12, 901–910. [Google Scholar] [CrossRef]
- Zhang, T.; Sun, X.; Qian, X.; Zhu, C.; Li, Q.; Li, H. PdSNF1, a sucrose non-fermenting protein kinase gene, is required for Penicillium digitatum conidiation and virulence. Appl. Microbiol. Biotechnol. 2013, 97, 5433–5445. [Google Scholar] [CrossRef]
- Gandía, M.; Harries, E.; Marcos, J.F. The myosin motor domain-containing chitin synthase PdChsVII is required for development, cell wall integrity and virulence in the citrus postharvest pathogen Penicillium digitatum. Fungal Gen. Biol. 2015, 67, 58–70. [Google Scholar] [CrossRef]
- Harries, E.; Gandía, M.; Carmona, L.; Marcos, J.F. The Penicillium digitatum protein O-mannosyl transferase Pmt2 is required for cell wall integrity, conidiogenesis, virulence and sensitivity to the antifungal peptide PAF26. Mol Plant Pathol 2015, 16, 748–761. [Google Scholar] [CrossRef]
- Vilanova, L.; López-Pérez, M.; Ballester, A.R.; Teixidó, N.; Usall, J.; Lara, I.; Viñas, I.; Torres, R.; González-Candelas, L. Differential contribution of the two major polygalacturonases from Penicillium digitatum to virulence towards citrus fruit. Int. J. Food Microbiol. 2018, 282, 16–23. [Google Scholar] [CrossRef]
- Zhang, T.; Qian, X.; Sun, X.; Li, H. The calcineurin-responsive transcription factor Crz1 is required for conidation, full virulence and DMI resistance in Penicillium digitatum. Microbiol Res. 2013, 168, 211–222. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Sun, X.; Xu, Q.; González-Candelas, L.; Li, H. The pH signaling transcription factor PacC is required for full virulence in Penicillium digitatum. Appl. Microbiol. Biotechnol. 2013, 97, 9087–9098. [Google Scholar] [CrossRef] [PubMed]
- Vilanova, L.; Teixidó, N.; Torres, R.; Usall, J.; Viñas, I.; Sánchez-Torres, P. Relevance of the transcription factor PdSte12 in Penicillium digitatum conidiation and virulence during citrus fruit infection. Int. J. Food Microbiol. 2016, 235, 93–102. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.; Sun, X.; Wang, M.; Gai, Y.; Chung, K.R.; Li, H. The citrus postharvest pathogen Penicillium digitatum depends on the PdMpkB kinase for developmental and virulence functions. Int. J. Food Microbiol. 2016, 236, 167–176. [Google Scholar] [CrossRef]
- De Ramón-Carbonell, M.; Sánchez-Torres, P. PdSlt2 Penicillium digitatum mitogen-activated-protein kinase controls sporulation and virulence during citrus fruit infection. Fungal Biol. 2017, 121, 1063–1074. [Google Scholar]








© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Ramón-Carbonell, M.; López-Pérez, M.; González-Candelas, L.; Sánchez-Torres, P. PdMFS1 Transporter Contributes to Penicilliun digitatum Fungicide Resistance and Fungal Virulence during Citrus Fruit Infection. J. Fungi 2019, 5, 100. https://doi.org/10.3390/jof5040100
de Ramón-Carbonell M, López-Pérez M, González-Candelas L, Sánchez-Torres P. PdMFS1 Transporter Contributes to Penicilliun digitatum Fungicide Resistance and Fungal Virulence during Citrus Fruit Infection. Journal of Fungi. 2019; 5(4):100. https://doi.org/10.3390/jof5040100
Chicago/Turabian Stylede Ramón-Carbonell, Marta, Mario López-Pérez, Luis González-Candelas, and Paloma Sánchez-Torres. 2019. "PdMFS1 Transporter Contributes to Penicilliun digitatum Fungicide Resistance and Fungal Virulence during Citrus Fruit Infection" Journal of Fungi 5, no. 4: 100. https://doi.org/10.3390/jof5040100
APA Stylede Ramón-Carbonell, M., López-Pérez, M., González-Candelas, L., & Sánchez-Torres, P. (2019). PdMFS1 Transporter Contributes to Penicilliun digitatum Fungicide Resistance and Fungal Virulence during Citrus Fruit Infection. Journal of Fungi, 5(4), 100. https://doi.org/10.3390/jof5040100