Antibiotic Activity of a Paraphaeosphaeria sporulosa-Produced Diketopiperazine against Salmonella enterica
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Isolation and Morphological Characterization
2.2. Molecular Characterization and Phylogenetic Analysis
2.3. Liquid Culture and Metabolite Production
2.4. Antibiotic Assay of Organic Extract
2.5. Extraction and Isolation of Secondary Metabolites
2.6. LC–MS Analysis
2.7. Antibiotic Assay on Fractions and Pure Compound
3. Results
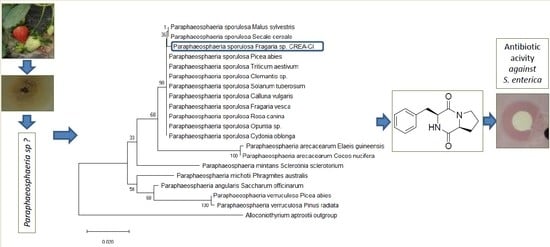
3.1. Morphological and Molecular Characterization of the Fungal Isolate
3.2. Isolation and Chemical Characterization of Bioactive Compound 1
3.3. Antibiotic Activity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Levy, S.B. Active efflux mechanisms for antimicrobial resistance. Antimicrob. Agents Chemother. 1992, 36, 695–703. [Google Scholar] [CrossRef] [Green Version]
- Jindal, B.A.K.; Pandya, M.K.; Khan, M.I.D. Antimicrobial resistance: A public health challenge. Med. J. Armed Forces India 2015, 71, 178–181. [Google Scholar] [CrossRef] [Green Version]
- Marquardt, R.R.; Li, S. Antimicrobial resistance in livestock: Advances and alternatives to antibiotics. Anim. Front. 2018, 8, 30–37. [Google Scholar] [CrossRef] [Green Version]
- Spellberg, B.; Gilbert, D.N. The future of antibiotics and resistance: A tribute to a career of leadership by John Bartlett. Clin. Infect. Dis. 2014, 59, S71–S75. [Google Scholar] [CrossRef] [Green Version]
- Hussain, C.; Ball, M.B.; McGwire, B.S. Multidrug-resistant bovine salmonellosis predisposing for severe human clostridial myonecrosis. Am. J. Case Rep. 2019, 20, 268–273. [Google Scholar] [CrossRef]
- Levy, S.B. The Antibiotic Paradox: How the Misuse of Antibiotics Destroys Their Curative Powers, 2nd ed.; Perseus Publishing: Cambridge, MA, USA, 2002. [Google Scholar]
- Fluckey, W.M.; Loneragan, G.H.; Warner, R.; Brashears, M.M. Antimicrobial drug resistance of Salmonella and Escherichia coli isolates from cattle feces, hides, and carcasses. J. Food Prot. 2007, 70, 551–556. [Google Scholar] [CrossRef]
- Kemal, J. A review on the public health importance of bovine salmonellosis. J. Veterinar. Sci. Technol. 2014, 5, 175. [Google Scholar] [CrossRef] [Green Version]
- Berdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [Green Version]
- Schulz, B.; Boyle, C.; Draeger, S.; Römmert, A.K.; Krohn, K. Endophytic fungi: A source of novel biologically active secondary metabolites. Mycol. Res. 2002, 106, 996–1004. [Google Scholar] [CrossRef]
- White, T.J.; Burns, T.; Lee, S.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications, 2nd ed.; Innis, M.A., Gelfand, D.H., Sninsky, H.H., White, T.J., Eds.; Academic Press: Cambridge, MA, USA, 1990. [Google Scholar]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 10 February 2015).
- Ritz, C.; Baty, F.; Streibig, J.C.; Gerhard, D. Dose-Response Analysis Using R. PLoS ONE 2015, 10, e0146021. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Welcome to the tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Verkley, G.J.M.; Dukik, K.; Renfurm, R.; Göker, M.; Stielow, J.B. Novel genera and species of coniothyrium-like fungi in Montagnulaceae (Ascomycota). Persoonia 2014, 32, 25–51. [Google Scholar] [CrossRef] [Green Version]
- Campbell, J.; Lin, Q.; Geske, G.D.; Blackwell, H.E. New and unexpected insights into the modulation of luxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol. 2009, 4, 1051–1059. [Google Scholar] [CrossRef]
- Jayatilake, G.S.; Thornton, M.P.; Leonard, A.C.; Grimwade, J.E.; Baker, B.J. Metabolites from an Antarctic sponge associated bacterium, Pseudomonas aeruginosa. J. Nat. Prod. 1996, 59, 293–296. [Google Scholar] [CrossRef]
- Borthwick, A.D. 2,5-Diketopiperazines: Synthesis, reactions, medicinal chemistry, and bioactive natural products. Chem. Rev. 2012, 112, 3641–3716. [Google Scholar] [CrossRef]
- Martins-Teixeira, M.; Carvalho, I. Diketopiperazines: Biological activity and synthesis. Tetrahedron 2007, 63, 9923. [Google Scholar] [CrossRef]
- Kumar, S.N.; Siji, J.V.; Nambisan, B.; Mohandas, C. Activity and synergistic antimicrobial activity between diketopiperazines against bacteria in vitro. Appl. Biochem. Biotechnol. 2012, 168, 2285–2296. [Google Scholar] [CrossRef]
- Guimarães, D.O.; Borges, W.S.; Vieira, N.J.; de Oliveira, L.F.; da Silva, C.H.; Lopes, N.P.; Dias, L.G.; Duran-Patron, R.; Collado, I.G.; Pupo, M.T. Diketopiperazines produced by endophytic fungi found in association with two Asteraceae species. Phytochemistry 2010, 71, 1423–1429. [Google Scholar] [CrossRef]
- Strobel, G.A. Endophytes as sources of bioactive products. Microb. Infect. 2011, 5, 535–544. [Google Scholar] [CrossRef]
- Nicoletti, R.; Vinale, F. Bioactive compounds from marine-derived Aspergillus, Penicillium, Talaromyces and Trichoderma species. Mar. Drugs 2018, 16, 408. [Google Scholar] [CrossRef] [Green Version]
- Cosoveanu, A.; Cabrera, R. Endophytic fungi in species of Artemisia. J. Fungi 2018, 4, 53. [Google Scholar] [CrossRef] [Green Version]
- Domsch, K.H.; Gams, W.; Anderson, T.H. Compendium of Soil Fungi, 2nd ed.; IHW-Verlag: Eching, Germany, 2007. [Google Scholar]
- Kothe, E.; Turnau, K. Mycorrhizosphere communication: Mycorrhizal fungi and endophytic fungus-plant interactions. Front. Microbiol. 2018, 9, 3015. [Google Scholar] [CrossRef] [Green Version]
- Liu, T.; Greenslade, A.; Yang, S. Levels of rhizome endophytic fungi fluctuate in Paris polyphylla var. yunnanensis as plants age. Plant Divers. 2017, 39, 60–64. [Google Scholar] [CrossRef]
- De Amorim, M.R. Bioprospection of Endophytic Fungi Paraphaeosphaeria sporulosa and Cochliobolus eragrostidis Isolated from Paepalanthus planifolius (Eriocaulaceae) and Epigenetic Study of Aspergillus terreus. Ph.D. Thesis, Universidade Estadual Paulista, Sao Paulo, Brazil, 2019. [Google Scholar]
- Zhao, C.; Fu, P.; Zhang, Y.; Liu, X.; Ren, F.; Che, Y. Sporulosol, a new ketal from the fungus Paracoconiothyrium sporulosum. Molecules 2018, 23, 1263. [Google Scholar] [CrossRef] [Green Version]
- Guiraud, P.; Steiman, R.; Seigle-Murandi, F.; Buarque de Gusmao, N. Antimicrobial and antitumor activities of mycosporulone. J. Nat. Prod. 1999, 62, 1222–1224. [Google Scholar] [CrossRef]
- Angulo, F.J.; Nargund, V.N.; Chiller, T.C. Evidence of an association between use of anti-microbial agents in food animals and antimicrobial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. 2004, 51, 374–379. [Google Scholar] [CrossRef]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carrieri, R.; Borriello, G.; Piccirillo, G.; Lahoz, E.; Sorrentino, R.; Cermola, M.; Censi, S.B.; Grauso, L.; Mangoni, A.; Vinale, F. Antibiotic Activity of a Paraphaeosphaeria sporulosa-Produced Diketopiperazine against Salmonella enterica. J. Fungi 2020, 6, 83. https://doi.org/10.3390/jof6020083
Carrieri R, Borriello G, Piccirillo G, Lahoz E, Sorrentino R, Cermola M, Censi SB, Grauso L, Mangoni A, Vinale F. Antibiotic Activity of a Paraphaeosphaeria sporulosa-Produced Diketopiperazine against Salmonella enterica. Journal of Fungi. 2020; 6(2):83. https://doi.org/10.3390/jof6020083
Chicago/Turabian StyleCarrieri, Raffaele, Giorgia Borriello, Giulio Piccirillo, Ernesto Lahoz, Roberto Sorrentino, Michele Cermola, Sergio Bolletti Censi, Laura Grauso, Alfonso Mangoni, and Francesco Vinale. 2020. "Antibiotic Activity of a Paraphaeosphaeria sporulosa-Produced Diketopiperazine against Salmonella enterica" Journal of Fungi 6, no. 2: 83. https://doi.org/10.3390/jof6020083
APA StyleCarrieri, R., Borriello, G., Piccirillo, G., Lahoz, E., Sorrentino, R., Cermola, M., Censi, S. B., Grauso, L., Mangoni, A., & Vinale, F. (2020). Antibiotic Activity of a Paraphaeosphaeria sporulosa-Produced Diketopiperazine against Salmonella enterica. Journal of Fungi, 6(2), 83. https://doi.org/10.3390/jof6020083