Exploring the Extracellular Macromolecular Composition of Crude Extracts of Penicillium rubens Strain 212 for Elucidation Its Mode of Action as a Biocontrol Agent
Abstract
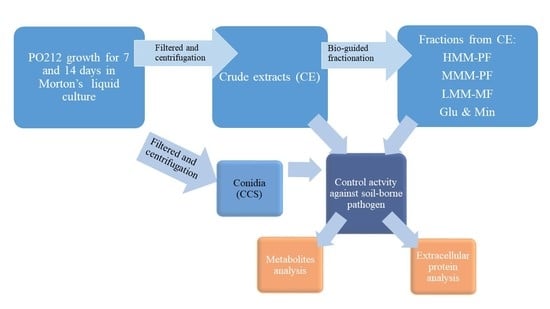
:1. Introduction
2. Materials and Methods
2.1. Microbial Cultures and Growth Conditions
2.2. Production of PO212 Conidia and Crude Extracts from Submerged Cultures
2.3. Evaluation of Control Activity of PO212 Conidia and Crude Extracts from Submerged Cultures Against FOL1A in Tomato Plants
2.4. Test of Antifungal Activity of PO212 Crude Extracts (CE7) Against FOL1A
2.5. Analysis of CE7 Low Molecular Mass Secondary Extracellular Metabolites
2.6. Bioassay-Guided Fractionation of PO212 Crude Extract (CE7)
2.6.1. Preparative Pressure-Driven Tangential-Flow Membrane UF
2.6.2. Preparative SPE
2.7. Evaluation of Disease Control Activiy of Purified Fractions from CE7
2.8. Multidimensional Liquid Chromatography Mass Spectrometry (nLC-MS) Identification of Proteins in Total and Fractionated CEs
2.9. Data Analysis
3. Results
3.1. Control Activity of PO212 Conidia and Crude Extracts from Submerged Cultures Against FOL1A in Tomato Plants
3.2. Test of the Antifungal Activity of PO212 Crude Extracts (CE7) Against FOL1A
3.3. Analysis of CE7 Low Molecular Mass Secondary Extracellular Metabolites
3.4. Bioassay-Guided Fractionation of PO212 Crude Extract (CE7)
3.5. Evaluation of Disease Control Activity of Purified Fractions from CE7
3.6. nLC-MS/MS Identification of Proteins in Total and Fractionated CEs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dean, R.; Van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edel-Hermann, V.; Lecomte, C. Current status of Fusarium oxysporum formae speciales and races. Phytopathology 2019, 109, 512–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kefalogianni, I.; Gkizi, D.; Pappa, E.; Dulaj, L.; Tjamos, S.E.; Chatzipavlidis, I. Combined use of biocontrol agents and zeolite as a management strategy against Fusarium and Verticillium wilt. BioControl 2017, 62, 139–150. [Google Scholar] [CrossRef]
- Aktar, W.; Sengupta, D.; Chowdhury, A. Impact of pesticides use in agriculture: Their benefits and hazards. Interdiscip. Toxicol. 2009, 2, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinale, F.; Sivasithamparam, K.; Ghisalberti, E.L.; Marra, R.; Woo, S.L.; Lorito, M. Trichoderma-plant-pathogen interactions. Soil Biol. Biochem. 2008, 40, 1–10. [Google Scholar] [CrossRef]
- Jiménez-Díaz, R.M.; Jiménez-Gasco, M.M. Integrated Management of Fusarium Wilt Diseases. In Control of Fusarium diseases; Alvés, F.M., Díez, J.J., Eds.; Research Signpost: Kerala, India, 2011; pp. 177–215. [Google Scholar]
- Bardin, M.; Ajouz, S.; Comby, M.; Lopez-Ferber, M.; Graillot, B.; Siegwart, M.; Nicot, P.C. Is the efficacy of biological control against plant diseases likely to be more durable than that of chemical pesticides? Front. Plant Sci. 2015, 6, 1–14. [Google Scholar] [CrossRef]
- Alvés-Sancho, F.M.; Díez, J.J. Biological control of Fusarium. In Control of Fusarium Diseases; Alves-Santos, F.M., Díez, J.J., Eds.; Research Signpost 37/661 (2) Fort P.O. Trivandum-695: Kerala, India, 2011; pp. 131–158. [Google Scholar]
- Bora, T.; Özaktan, H.; Göre, E.; Aslan, E. Biological control of Fusarium oxysporum f. sp. melonis by wettable powder formulations of the two strains of Pseudomonas putida. J. Phytopathol. 2004, 152, 471–475. [Google Scholar] [CrossRef]
- van Lenteren, J.C.; Bolckmans, K.; Köhl, J.; Ravensberg, W.J.; Urbaneja, A. Biological control using invertebrates and microorganisms: Plenty of new opportunities. Biol. Control 2018, 63, 39–59. [Google Scholar] [CrossRef] [Green Version]
- Ghorbanpour, M.; Omidvari, M.; Abbaszadeh-Dahaji, P.; Omidvar, R.; Kariman, K. Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 2018, 117, 147–157. [Google Scholar] [CrossRef]
- Alabouvette, C.; Olivain, C.; Migheli, Q.; Steinberg, C. Microbiological control of soil-borne phytopathogenic fungi with special emphasis on wilt-inducing Fusarium oxysporum. New Phytol. 2009, 184, 529–544. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 1–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massart, S.; Martinez-Medina, M.; Haissam, J.M. Biological control in the microbiome era : Challenges and opportunities. Biol. Control 2015, 89, 98–108. [Google Scholar] [CrossRef]
- Woo, S.L.; Scala, F.; Ruocco, M.; Lorito, M. The molecular biology of the interactions between Trichoderma spp., phytopathogenic fungi, and plants. Phytopathology 2006, 96, 181–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hanson, L.E.; Howell, C.R. Elicitors of plant defense responses from biocontrol strains of Trichoderma virens. Phytopathology 2004, 94, 171–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarino, M.; De Cal, A.; Melgarejo, P.; Larena, I.; Espeso, E.A. The development of genetic and molecular markers to register and commercialize Penicillium rubens (formerly Penicillium oxalicum) strain 212 as a biocontrol agent. Microb. Biotechnol. 2016, 9, 89–99. [Google Scholar] [CrossRef]
- De Cal, A.; Pascual, S.; Larena, I.; Melgarejo, P. Biological control of Fusarium oxysporum f. sp. lycopersici. Plant Pathol. 1995, 44, 909–917. [Google Scholar] [CrossRef]
- De Cal, A.; García-Lepe, R.; Pascual, S.; Melgarejo, P. Effects of timing and method of application of Penicillium oxalicum on efficacy and duration of control of Fusarium wilt of tomato. Plant Pathol. 1999, 48, 260–266. [Google Scholar] [CrossRef]
- De Cal, A.; Redondo, C.; Sztejnberg, A.; Melgarejo, P. Biocontrol of powdery mildew by Penicillium oxalicum in open-field nurseries of strawberries. Biol. Control 2008, 47, 103–107. [Google Scholar] [CrossRef]
- De Cal, A.; Sztejnberg, A.; Sabuquillo, P.; Melgarejo, P. Management Fusarium wilt on melon and watermelon by Penicillium oxalicum. Biol. Control 2009, 51, 480–486. [Google Scholar] [CrossRef]
- Larena, I.; Sabuquillo, P.; Melgarejo, P.; De Cal, A. Biocontrol of Fusarium and Verticillium wilt of tomato by Penicillium oxalicum under greenhouse and field conditions. J. Phytopathol. 2003, 151. [Google Scholar] [CrossRef]
- Sabuquillo, P.; Cal, A.; Melgarejo, P. Biocontrol of tomato wilt by Penicillium oxalicum formulations in different crop conditions. Biol. Control 2006, 37, 256–265. [Google Scholar] [CrossRef]
- Martínez-Beringola, M.L.; Salto, T.; Vázquez, G.; Larena, I.; Melgarejo, P.; De Cal, A. Penicillium oxalicum reduces the number of cysts and juveniles of potato cyst nematodes. J. Appl. Microbiol. 2013, 115. [Google Scholar] [CrossRef] [PubMed]
- De Cal, A.; Pascual, S.; Melgarejo, P. Involvement of resistance induction by Penicillum oxalicum in the biocontrol of tomato wilt. Plant Pathol. 1997, 46, 72–79. [Google Scholar] [CrossRef]
- De Cal, A.; Garcia-Lepe, R.; Melgarejo, P. Induced resistance by Penicillium oxalicum against Fusarium oxysporum f. sp. lycopersici: Histological studies of infected and induced tomato stems. Phytopathology 2000, 90, 260–268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- García-Lepe, R.; Rodriguez, P.; De Cal, A.; García-Olmedo, F.; Melgarejo, P. Induced resistance against Fusarium wilt of tomato by Penicillium oxalicum is not associated to pathogenesis-related proteins. IOBC Bull. 1997, 21, 123–127. [Google Scholar]
- Pascual, S.; Melgarejo, P.; Magan, N. Induction of submerged conidiation of the biocontrol agent Penicillium oxalicum. Appl. Microbiol. Biotechnol. 1997, 48, 389–392. [Google Scholar] [CrossRef]
- Larena, I.; Melgarejo, P.; De Cal, A. Drying of conidia of Penicillium oxalicum, a biological control agent against Fusarium wilt of tomato. J. Phytopathol. 2003, 151. [Google Scholar] [CrossRef]
- De Cal, A.; Mateo-Sagasta, E.; Melgarejo, P. Antifungal substances produced by Penicillium frequentans and their relationship to the biocontrol of Monilinia laxa. Phytopathology 1988, 78, 888–893. [Google Scholar] [CrossRef] [Green Version]
- Larena, I.; Melgarejo, P.; De Cal, A. Production, survival, and evaluation of solid-substrate inocula of Penicillium oxalicum, a biocontrol agent against Fusarium wilt of tomato. Phytopathology 2002, 92, 863–869. [Google Scholar] [CrossRef]
- Pascual, S.; De Cal, A.; Magan, N.; Melgarejo, P. Surface hydrophobicity, viability and efficacy in biological control of Penicillium oxalicum spores produced in aerial and submerged culture. J. Appl. Microbiol. 2000, 89, 847–853. [Google Scholar] [CrossRef] [Green Version]
- Larena, I.; Melgarejo, P. Development of a method for detection of the biocontrol agent Penicillium oxalicum strain 212 by combining PCR and a selective medium. Plant Dis. 2009, 93, 919–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoagland, D.R.; Arnon, D.I. The Water-Culture Method for Growing Plants without Soil, 2nd ed.; Arnon, D.I., Ed.; College of Agriculture, University of California: Berkeley, CA, USA, 1950; Volume 347. [Google Scholar]
- Campbell, C.L.; Madden, L.V. Introduction to Plant Disease Epidemiology; John Wiley & Sons: New York, NY, USA, 1990. [Google Scholar]
- De Cal, A.; Larena, I.; Guijarro, B.; Melgarejo, P. Mass production of conidia of Penicillium frequentans, a biocontrol agent against brown rot of stone fruits. Biocontrol Sci. Technol. 2002, 12, 715–725. [Google Scholar] [CrossRef]
- Gómez Ramos, M.J.; Lozano, A.; Fernández-Alba, A.R. High-resolution mass spectrometry with data independent acquisition for the comprehensive non-targeted analysis of migrating chemicals coming from multilayer plastic packaging materials used for fruit purée and juice. Talanta 2019, 191, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Silván, J.M.; Pinto-Bustillos, M.A.; Vásquez-Ponce, P.; Prodanov, M.; Martinez-Rodriguez, A.J. Olive mill wastewater as a potential source of antibacterial and anti-inflammatory compounds against the food-borne pathogen Campylobacter. Innov. Food Sci. Emerg. Technol. 2019, 51, 177–185. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Ortiz, P.; Espeso, E.A. Phospho-regulation and nucleocytoplasmic trafficking of CrzA in response to calcium and alkaline-pH stress in Aspergillus nidulans. Mol. Microbiol. 2013, 89, 532–551. [Google Scholar] [CrossRef] [PubMed]
- Sambrook, J.; Russell, D. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: New York, NY, USA, 2001; ISBN 978-087969577-4. [Google Scholar]
- Manoli, M.; Espeso, E.A. Modulation of calcineurin activity in Aspergillus nidulans: The roles of high magnesium concentrations and of transcriptional factor CrzA. Mol. Microbiol. 2019, 111, 1283–1301. [Google Scholar] [CrossRef] [Green Version]
- Van den Berg, M.; Albang, R.; Albermann, K.; Badger, J.; Daran, J.; Driessen, A.; Garcia-Estrada, C.; Fedorova, N.; Harris, D.; Heijne, W.; et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat. Biotechnol. 2008, 26, 1161–1168. [Google Scholar] [CrossRef] [Green Version]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Fleming’s penicillin producing strain is not Penicillium chrysogenum but P. rubens. IMA Fungus 2011, 2, 87–95. [Google Scholar] [CrossRef] [Green Version]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018, 47, 309–314. [Google Scholar] [CrossRef] [Green Version]
- Almagro Armenteros, J.J.; Tsirigos, K.D.; Sønderby, C.K.; Petersen, T.N.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 2019, 37, 420–423. [Google Scholar] [CrossRef] [PubMed]
- Snedecor, G.W.; Cochran, W.G. Statistical Methods; University Press: Ames, IA, USA, 1980. [Google Scholar]
- Díez, B.; Marcos, A.T.; Rodríguez, M.; Luis de la Fuente, J.; Barredo, J.L. Structural and phylogenetic analysis of the γ-actin encoding gene from the penicillin-producing fungus Penicillium chrysogenum. Curr. Microbiol. 2001, 42, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Basco, M.J.; Bisen, K.; Keswani, C.; Singh, H.B. Biological management of Fusarium wilt of tomato using biofortified vermicompost. Mycosphere 2017, 8, 467–483. [Google Scholar] [CrossRef]
- Mishra, J.; Tewari, S.; Singh, S.; Arora, N. Biopesticides where we stand? In Plant Microbe Symbiosis: Applied Facets; Arora, N., Ed.; Springer: New Delhi, India, 2015; pp. 37–75. [Google Scholar]
- Sarma, B.K.; Yadav, S.K.; Patel, J.S.; Singh, H.B. Molecular mechanisms of interactions of Trichoderma with other fungal species. Open Mycol. J. 2014, 8, 140–147. [Google Scholar] [CrossRef] [Green Version]
- Carrizales, V.; Jaffe, W. Solid state fermentation an appropriate biotechnology for developing countries. Interscience 1986, 11, 9–15. [Google Scholar]
- Raaijmakers, J.; Leeman, M.; Van Oorschot, M.; Van der Sluis, I.; Schippers, B.; Bakker, P. Dose-response relationships in biological control of fusarium wilt of radish by Pseudomonas spp. Phytopathology 1995, 85, 1075–1081. [Google Scholar] [CrossRef]
- Muñoz, G.A.; Agosin, E.; Cotoras, M.; San Martin, R.; Volpe, D. Comparison of aerial and submerged spore properties for Trichoderma harzianum. FEMS Microbiol. Lett. 1995, 125, 65–70. [Google Scholar] [CrossRef]
- Jackson, M.; Schisler, D. The composition and attributes of Colletotrichum truncatum spores are altered by the nutritional environment. Appl. Environ. Microbiol. 1992, 58, 2260–2265. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Sun, M.H.; Liu, X.Z.; Che, Y.S. Effects of carbon concentration and carbon to nitrogen ratio on the growth and sporulation of several biocontrol fungi. Mycol. Res. 2007, 111, 87–92. [Google Scholar] [CrossRef]
- Espeso, E.A.; Villarino, M.; Carreras, M.; Alonso-Guirado, L.; Alonso, J.M.; Melgarejo, P.; Larena, I. Altered nitrogen metabolism in biocontrol strains of Penicillium rubens. Fungal Genet. Biol. 2019, 132. [Google Scholar] [CrossRef]
- Thuerig, B.; Felix, G.; Binder, A.; Boller, T.; Tamm, L. An extract of Penicillium chrysogenum elicits early defense-related responses and induces resistance in Arabidopsis thaliana independently of known signalling pathways. Physiol. Mol. Plant Pathol. 2006, 67, 180–193. [Google Scholar] [CrossRef]
- Tamm, L.; Thürig, B.; Fliessbach, A.; Goltlieb, A.E.; Karavani, S.; Cohen, Y. Elicitors and soil management to induce resistance against fungal plant diseases. NJAS Wageningen J. Life Sci. 2011, 58, 131–137. [Google Scholar] [CrossRef]
- Hu, X.; Qin, L.; Roberts, D.P.; Lakshman, D.K.; Gong, Y.; Maul, J.E.; Xie, L.; Yu, C.; Li, Y.; Hu, L.; et al. Characterization of mechanisms underlying degradation of sclerotia of Sclerotinia sclerotiorum by Aspergillus aculeatus Asp-4 using a combined qRT-PCR and proteomic approach. BMC Genom. 2017, 18, 674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Chen, L.; Sun, Y.; Zhao, L.; Zheng, X.; Yang, Q.; Zhang, X. Investigating proteome and transcriptome defense response of apples induced by Yarrowia lipolytica. Mol. Plant-Microbe Interact. 2017, 30, 301–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massart, S.; Perazzolli, M.; Haı, P.M. Impact of the omic technologies for understanding the modes of action of biological control agents against plant pathogens. Biol. Control. 2015, 725–746. [Google Scholar] [CrossRef]
- Villarino, M.; Espeso, E.A.; Melgarejo, P.; Larena, I. Transformation of Penicillium rubens 212 and expression of GFP and DsRED coding genes for visualization of plant-biocontrol agent interaction. Front. Microbiol. 2018. [Google Scholar] [CrossRef] [Green Version]
- Druzhinina, I.S.; Komoń-Zelazowska, M.; Ismaiel, A.; Jaklitsch, W.; Mullaw, T.; Samuels, G.J.; Kubicek, C.P. Molecular phylogeny and species delimitation in the section Longibrachiatum of Trichoderma. Fungal Genet. Biol. 2012, 49, 358–368. [Google Scholar] [CrossRef] [Green Version]
- Lima, F.B.; Félix, C.; Osório, N.; Alves, A.; Vitorino, R.; Domingues, P.; da Silva Ribeiro, R.T.; Esteves, A.C. Trichoderma harzianum T1A constitutively secretes proteins involved in the biological control of Guignardia citricarpa. Biol. Control 2017, 106, 99–109. [Google Scholar] [CrossRef]
- Atanasova, L.; Le Crom, S.; Gruber, S.; Coulpier, F.; Seidl-Seiboth, V.; Kubicek, C.P.; Druzhinina, I.S. Comparative transcriptomics reveals different strategies of Trichoderma mycoparasitism. BMC Genom. 2013, 14, 121. [Google Scholar] [CrossRef] [Green Version]
- De Marco, J.L.; Felix, C.R. Characterization of a protease produced by a Trichoderma harzianum isolate which controls cocoa plant witches’ broom disease. BMC Biochem. 2002, 3, 3. [Google Scholar] [CrossRef]
- Grinyer, J.; Hunt, S.; McKay, M.; Herbert, B.R.; Nevalainen, H. Proteomic response of the biological control fungus Trichoderma atroviride to growth on the cell walls of Rhizoctonia solani. Curr. Genet. 2005, 47, 381–388. [Google Scholar] [CrossRef]
- Pozo, M.J.; Baek, J.M.; García, J.M.; Kenerley, C.M. Functional analysis of tvsp1, a serine protease-encoding gene in the biocontrol agent Trichoderma virens. Fungal Genet. Biol. 2004, 41, 336–348. [Google Scholar] [CrossRef]
- Meijer, H.J.G.; Mancuso, F.M.; Espadas, G.; Seidl, M.F.; Chiva, C.; Govers, F.; Sabidó, E. Profiling the secretome and extracellular proteome of the potato late blight pathogen Phytophthora infestans. Mol. Cell. Proteom. 2014, 13, 2101–2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nogueira-Lopez, G.; Greenwood, D.R.; Middleditch, M.; Winefield, C.; Eaton, C.; Steyaert, J.M.; Mendoza-Mendoza, A. The apoplastic secretome of Trichoderma virens during interaction with maize roots shows an inhibition of plant defence and scavenging oxidative stress secreted proteins. Front. Plant Sci. 2018, 9, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Vivek-Ananth, R.P.; Mohanraj, K.; Vandanashree, M.; Jhingran, A.; Craig, J.P.; Samal, A. Comparative systems analysis of the secretome of the opportunistic pathogen Aspergillus fumigatus and other Aspergillus species. Sci. Rep. 2018, 8, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Friel, D.; Pessoa, N.M.G.; Vandenbol, M.; Jijakli, M.H. Separate and combined disruptions of two exo-β-1,3-glucanase genes decrease the efficiency of Pichia anomala (strain K) biocontrol against Botrytis cinerea on apple. Mol. Plant-Microbe Interact. 2007, 20, 371–379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coda, R.; Cassone, A.; Rizzello, C.G.; Nionelli, L.; Cardinali, G.; Gobbetti, M. Antifungal activity of Wickerhamomyces anomalus and Lactobacillus plantarum during sourdough fermentation: Identification of novel compounds and long-term effect during storage of wheat bread. Appl. Environ. Microbiol. 2011, 77, 3484–3492. [Google Scholar] [CrossRef] [Green Version]
- Jijakli, M.H. Pichia anomala in biocontrol for apples: 20 years of fundamental research and practical applications. Anton. Leeuw. Int. J. G 2011, 99, 93e105. [Google Scholar]
- Ferre, F.S.; Santamarina, M.P. Efficacy of Trichoderma harzianum in suppression of Fusarium culmorum. Ann. Microbiol. 2010, 60, 335–340. [Google Scholar] [CrossRef]
- Cantín, A.; Moya, P.; Castillo, M.A.; Primo, J.; Miranda, M.; Primo-Yúfera, E. Isolation and synthesis of N-(2.Methyl-3-oxodec-8-enoyl)-2-pyrroline and 2-(Hept-5-enyl)-3-methyl-4-oxo-6,7,8,8a-tetrahydro-4H-pyrrolo[2,1-b]-1,3-oxazine—Two new fungal metabolites with in vivo anti-juvenile hormone and insecticidal activity. Eur. J. Org. Chem. 1999, 1, 221–226. [Google Scholar] [CrossRef]
- Cantín, Á.; Moya, P.; Miranda, M.A.; Primo, J.; Primo-Ýfera, E. Isolation of N-(2-Methyl-3-oxodecanoyl)pyrrole and N-(2-Methyl-3-oxodec-8-enoyl)pyrrole, two new natural products from Penicillium brevicompactum, and synthesis of analogues with insecticidal and fungicidal activity. J. Agric. Food Chem. 1998, 46, 4748–4753. [Google Scholar] [CrossRef]
- Bhardwaj, V.; Gumber, D.; Abbot, V.; Dhiman, S.; Sharma, P. Pyrrole: A resourceful small molecule in key medicinal hetero-aromatics. RSC Adv. 2015, 5, 15233–15266. [Google Scholar] [CrossRef]
- Yadav, R.N.; Mahtab Rashid, M.; Zaidi, N.W.; Kumar, R.; Singh, H.B. Secondary metabolites of Metarhizium spp. and Verticillium spp. and their agricultural applications. In Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms; Singh, H., Keswani, C., Reddy, M., Sansinenea, E., García-Estrada, C., Eds.; Springer: Singapore, 2019. [Google Scholar]
- Veselinović, A.M.; Nikolić, G.M. Influence of Zn(II) ion on the autoxidation of pyrogallol and gallic acid in weakly acidic aqueous solutions. Acta Fac. Medicae Naissensis 2015, 32, 127–135. [Google Scholar] [CrossRef] [Green Version]
- Guzmán-Chávez, F.; Salo, O.; Nygård, Y.; Lankhorst, P.P.; Bovenberg, R.A.L.; Driessen, A.J.M. Mechanism and regulation of sorbicillin biosynthesis by Penicillium chrysogenum. Microb. Biotechnol. 2017, 10, 958–968. [Google Scholar] [CrossRef]






| Treatment | Description | Application Dose | Assay |
|---|---|---|---|
| PO212 1 | PO212 conidia after growing into solid fermentation and dried in fluid-drier | 1 × 107 conidia g−1 substrate | A1, A2, A3, A4 |
| aPO212 1,2 | Autoclaved PO212 conidial suspension | 1 × 107 conidia g−1 substrate | A1 |
| SCC7 1 | PO212 conidia after growing into Morton’s medium for 7 days | 1 × 107 conidia g−1 substrate | A1 |
| SCC14 1 | PO212 conidia after growing into Morton’s medium for 14 days | 1 × 107 conidia g−1 substrate | A1 |
| CE7 | PO212 crude extract after growing into Morton’s medium for 7 days | 60 mL kg−1 substrate | A1, A2, A3, A4 |
| CE14 | PO212 crude extracts after growing into Morton’s medium for 14 days | 60 mL kg−1 substrate | A1 |
| aCE7 2 | CE7 was autoclaved | 60 mL kg−1 substrate | A1 |
| aCE14 2 | CE14 was autoclaved | 60 mL kg−1 substrate | A1 |
| CE7 1:1 | CE7 was mixed with SDW in a ratio of 1:1 (v/v) | 60 mL kg−1 substrate | A2 |
| CE7 1:10 | CE7 was mixed with SDW in a ratio of 1:10 (v/v) | 60 mL kg−1 substrate | A2 |
| CE7 1:50 | CE7 was mixed with SDW in a ratio of 1:50 (v/v) | 60 mL kg−1 substrate | A2 |
| d-CE7 | CE7 was treated with proteinase K and DB at 50 °C for 1 h | 60 mL kg−1 substrate | A3 |
| 20f-CE7 | CE7 was kept at −20 °C for 24 h and then it was thawed under agitation to RT3 | 60 mL kg−1 substrate | A4 |
| 80f-CE7 | CE7 was kept at −80 °C for 24h and then it was thawed under agitation to RT3 | 60 mL kg−1 substrate | A4 |
| DB | Digestion buffer 5 × (250 mM Tris-HCl pH 7.5 and 25 mM CaCl2) | 60 mL kg−1 substrate | A3 |
| Morton | Morton’s B medium | 60 mL kg−1 substrate | A1, A2, A3, A4 |
| C | Control with SDW | 60 mL kg−1 substrate | A1, A2, A3, A4 |
| Treatment | Disease Severity (%) | Disease Reduction (%) | AUDPC |
|---|---|---|---|
| PO212 | 6.9 d | 79.1 | 449.2 c |
| aPO212 | 33.9 ab | 0 | 802.5 ab |
| CE7 | 12.7 d | 63.6 | 432.8 c |
| aCE7 | 26.4 c | 20.0 | 821.3 ab |
| CE14 | 17.1 d | 48.2 | 551.3 c |
| aCE14 | 25.2 c | 23.6 | 748.0 b |
| SCC7 | 31.8 b | 3.6 | 805.5 ab |
| SCC14 | 37.5 a | 0 | 1015.0 a |
| Morton | 25.6 c | 22.4 | 824.5 ab |
| C1 | 33.0 ab | - | 782.5 ab |
| MSE 1 | 325.6 | 14852.8 |
| Treatment | Nutritive Solution Consumption (mL Plant−1 Day−1) | Leaves Number Plant−1 | Fresh Stem Weight (g Plant−1) | Root Weight (g Plant−1) |
|---|---|---|---|---|
| PO212 | 4.9 | 8.9 a | 9.5 a | 6.5 a |
| aPO212 | 4.9 | 7.8 b | 8.2 b | 5.5 cd |
| CE7 | 5.0 | 9.0 a | 9.7 a | 5.7 c |
| aCE7 | 4.8 | 9.1 a | 8.0 b | 5.4 cde |
| CE14 | 4.9 | 8.3 ab | 8.1 b | 5.7 c |
| aCE14 | 4.7 | 8.3 ab | 8.1 b | 5.4 cde |
| SCC7 | 5.0 | 8.3 ab | 8.7 b | 5.2 de |
| SCC14 | 4.7 | 8.3 ab | 7.8 b | 5.1 e |
| Morton | 4.7 | 8.0 b | 7.8 b | 5.0 e |
| C2 | 3.0 | 7.9 b | 7.8 b | 6.2 b |
| MSE 1 | 3.6 NS | 1.2 | 5.7 | 2.0 |
| Treatment | Disease Severity (%) | Disease Reduction (%) | Disease Incidence (%) | AUDPC |
|---|---|---|---|---|
| PO212 | 33.1 c | 32.2 | 40.0 b | 636.8 c |
| CE7 | 33.4 c | 31.6 | 62.5 a | 629.3 c |
| CE7 1:1 | 41.7 b | 14.5 | 64.1 a | 698.1 bc |
| CE7 1:10 | 40.5 b | 17.0 | 70.0 a | 792.7 bc |
| CE7 1:50 | 51.6 a | 0 | 68.8 a | 1005.0 a |
| Morton 1 | 53.6 a | 0 | 60.0 a | 1027.5 a |
| C1 2 | 48.8 a | - | 65.0 a | 808.9 b |
| MSE 3 | 143.4 | 2043.7 | 0.1 |
| Concentration (%) | Micelial Growth (mm day −1) | Sporulation (Conidia Number Plate−1) | Germ Tubes Length (µm) | Germination (%) |
|---|---|---|---|---|
| 0 (Control) | 1.3 | 5.7 × 108 (20.2) a | 11.7 d | 46.8 (27.9) c |
| 2.5 | ND | ND | 15.9 c | 74.5 (48.2) b |
| 12.5 | ND | ND | 16.1 b | 75.5 (49.1) ab |
| 25.0 | 1.3 | 4.1 × 108 (19.8) a | 16.8 ab | 77.8 (51.0) ab |
| 50.0 | 1.3 | 1.5 × 108 (18.8) b | 17.6 a | 78.5 (51.7) a |
| 80.0 | 1.3 | 2.2 × 107 (16.7) c | ND | ND |
| MSE 1 | 1.4 × 10−4 NS | 2.9 × 1017 (11.9) | 26.7 | 299.1 |
| Treatment | Disease Severity (%) | Disease Reduction (%) | AUDPC | Disease Incidence (%) | Leaf Number Plant−1 | Stem Weight (g Plant−1) | Root Weight (g Plant−1) | Nutritive Solution Consumption (mL Plant−1 Day−1) |
|---|---|---|---|---|---|---|---|---|
| PO212 * | 11.1 f | 74.3 | 229.4 b | 35.0 bc | 6.3 b | 1.0 e | 1.2 c | 1.7 d |
| PO212 | 0.0 g | 0.0 c | 0.0 c | 8.2 a | 3.3 a | 1.9 a | 5.1 b | |
| CE7 * | 16.3 ef | 62.3 | 250.3 b | 50.0 b | 6.4 b | 0.6 efg | 0.8 e | 1.5 d |
| CE7 | 0.0 g | 0.0 c | 0.0 c | 7.0 b | 2.7 b | 1.7 b | 3.9 bc | |
| HMM-PF * | 19.3 de | 55.3 | 225.0 b | 25.0 bc | 6.1 b | 0.7 efg | 0.7 e | 1.5 d |
| HMM-PF | 0.0 g | 0.0 c | 0.0 c | 6.2 b | 2.4 c | 1.3 c | 4.3 bc | |
| MMM-PF * | 10.9 f | 74.8 | 282.6 b | 50.0 b | 4.9 cd | 0.7 ef | 0.6 ef | 1.2 d |
| MMM-PF | 0.0 g | 0.0 c | 0.0 c | 6.9 b | 2.1 c | 1.1 cd | 6.8 a | |
| Glu&Min * | 25.4 cd | 41.2 | 326.5 b | 85.0 a | 4.6 d | 0.4 fgh | 0.4 f | 0.6 d |
| Glu&Min | 0.0 g | 0.0 c | 0.0 c | 6.7 b | 1.5 d | 0.8 e | 4.3 bc | |
| LMM-MF * | 30.3 c | 29.9 | 252.4 b | 40.0 b | 3.9 d | 0.3 gh | 0.4 f | 0.7 d |
| LMM-MF | 0.0 g | 0.0 c | 0.0 c | 5.7 bc | 1.5 d | 0.9 de | 3.0 c | |
| Morton B * | 49.7 a | 0 | 471.5 a | 83.3 a | 6.2 b | 0.5 fgh | 0.6 ef | 0.7 d |
| Morton B | 0.0 g | 0.0 c | 0.0 c | 6.1 b | 1.7 d | 1.1 c | 3.6 c | |
| C1 * | 43.2 b | 521.3 a | 85.0 a | 4.0 d | 0.2 h | 0.4 f | 0.2 d | |
| C2 | 0.0 g | 0.0 c | 0.0 c | 6.9 b | 2.0 c | 1.1 cd | 3.1 c | |
| MSE 1 | 24.18 | 4092.89 | 355.03 | 0.45 | 0.06 | 0.02 | 0.61 |
| Treatment | Disease Severity (%) | Disease Reduction (%) | AUDPC | Disease Incidence (%) | Leaf Number Plant−1 | Stem Weight (g Plant−1) | Root Weight (g Plant−1) | Nutritive Solution Consumption (mL Plant−1 Day−1) |
|---|---|---|---|---|---|---|---|---|
| PO212 | 11.1 c | 74.3 | 229.4 (5.4) b | 35.0 bc | 6.3 a | 1.0 b | 1.2 a | 1.7 b |
| CE7 | 16.3 bc | 62.3 | 250.3 (5.5) b | 50.0 ab | 6.4 a | 0.6 c | 0.8 c | 1.5 b |
| CE7L | 13.7 c | 68.3 | 198.3 (5.3) b | 50.0 ab | 4.0 c | 0.3 de | 0.4 d | 0.7 bc |
| HMM-PF | 19.3 bc | 55.3 | 225.0 (5.4) b | 25.0 bc | 6.1 a | 0.7 c | 0.7 c | 1.5 b |
| HMM-PF/L | 24.5 b | 43.3 | 246.9 (5.5) b | 45.0 ab | 5.2 b | 0.5 cd | 0.4 d | 1.0 bc |
| MMM-PF | 10.9 c | 74.8 | 282.6 (5.6) b | 50.0 ab | 4.9 bc | 0.7 c | 0.6 c | 1.2 b |
| MMM-PF/L | 11.3 c | 73.8 | 286.8 (5.6) b | 40.0 bc | 4.7 bc | 0.7 c | 0.6 c | 1.3 b |
| Morton B | 49.7 a | 0 | 471.5 (6.2) a | 83.3 a | 6.2 a | 0.5 cd | 0.6 c | 0.7 bc |
| C1 | 43.2 a | 0 | 521.3 (6.2) a | 85.0 a | 4.0 a | 0.2 e | 0.4 d | 0.2 c |
| C2 | 0.0 d | 0.0 (0.0) c | 0.0 c | 6.9 a | 2.0 a | 1.1 b | 3.1 a | |
| MSE 1 | 39.98 | 3642.67 | 499.30 | 0.26 | 0.02 | 0.01 | 0.30 |
| Proteins | PO212 Gene ID | Function | MM 1 (kDa) | pI 1 |
|---|---|---|---|---|
| With Signal Peptide | ||||
| Serine peptidase-type carboxypeptidase family 10 | g9920 | Proteolysis | 57.6 | 5.4 |
| Serine peptidase-type carboxypeptidase family 10 | g9786 | Proteolysis | 61.4 | 4.9 |
| Serine alkaline protease family 8 | g5776 | Endopeptidase activity | 40.3 | 6.2 |
| Flavin adenine dinucleotide binding | g7429 | Oxidoreductase activity | 61.6 | 8.5 |
| Flavin adenine dinucleotide binding | g9078 | Oxidoreductase activity | 60.1 | 8.3 |
| Flavin adenine dinucleotide binding | g6703 | Oxidoreductase activity | 48.4 | 4.6 |
| GH family 17 | g4058 | Metabolic process | 45.6 | 4.8 |
| Ion calcio binding–1,2-α-mannosidase activity family 47 | g3615 | Metabolic process | 56.9 | 5.1 |
| Glucan endo-1,6-β-glucosidase family 5 | g5409 | Carbohydrate metabolic process | 67.5 | 4.5 |
| Hydrolase O-glycosyl compounds family 3 | g9789 | Carbohydrate metabolic process | 52.0 | 5.2 |
| Glucan 1,4-α-glucosidase family 15 | g8238 | Carbohydrate metabolic process | 56.4 | 5.2 |
| GH family 32 | g3404 | Carbohydrate metabolic process | 77.5 | 4.9 |
| 1,3-β-glucanosil transferase family 72 | g9967 | Carbohydrate metabolic process | 36.0 | 4.9 |
| GH family 16 | g9585 | Carbohydrate metabolic process | 35.8 | 6.7 |
| GH family 43 | g1771 | Carbohydrate metabolic process | 41.7 | 5.7 |
| Uncharacterized protein | g4235 | Uncharacterized protein | 40.9 | 4.8 |
| Uncharacterized protein | g9424 | Uncharacterized protein | 43.3 | 5.5 |
| Others | g37 | Transmembrane carrier | 61.8 | 4.8 |
| Lacking Signal Peptide | ||||
| GAPDH activity, NAD and NADP binding | g5740 | Oxidoreductase activity | 36.0 | 6.7 |
| GH family 32 | g4776 | Carbohydrate metabolic process | 62.5 | 5.3 |
| ATP binding | g6856 | Gamma-actin | 59.5 | 8.9 |
| ATP binding | g7817 | Hydrolase and proton-transporting ATP synthase activity | 61.8 | 5.7 |
| ATP binding | g3421 | Chaperonin | 49.5 | 4.7 |
| Others | g468 | Putative cation/Cl− transporter | 130.9 | 6.6 |
| Others | g7400 | RNA binding | 82.3 | 8.3 |
| Others | g461 | Transmembrane transporter activity | 139.7 | 6.8 |
| Others | g5657 | Transcription initiation from RNA polymerase III promoter | 69.2 | 5.4 |
| Uncharacterized protein | g6156 | Uncharacterized protein | 30.7 | 10.4 |
| Uncharacterized protein | g7827 | Uncharacterized protein | 40.3 | 6.6 |
| Uncharacterized protein | g6729 | Uncharacterized protein | 129.1 | 6.7 |
| Proteins | PO212 Gene ID | Function | MM 1 (kDa) | pI 1 |
|---|---|---|---|---|
| With Signal Peptide | ||||
| Serine peptidase-type carboxypeptidase family 10 | g9920 | Proteolysis | 61.8 | 4.8 |
| Serine alkaline protease family 8 | g5776 | Endopeptidase activity | 40.3 | 6.2 |
| Flavin adenine dinucleotide binding | g9251 | Oxidoreductase activity | 50.3 | 5.5 |
| Flavin adenine dinucleotide binding | g9078 | Oxidoreductase activity | 61.6 | 8.5 |
| Flavin adenine dinucleotide binding | g6703 | Oxidoreductase activity | 60.1 | 8.3 |
| GH family 17 | g4058 | Metabolism | 48.4 | 4.6 |
| Ion calcio binding–1,2-α-mannosidase activity family 47 | g3615 | Metabolism | 56.4 | 5.2 |
| Hydrolase O-glycosyl compounds family 3 | g9789 | Carbohydrate metabolic process | 77.5 | 4.9 |
| Glucan 1,4-α-glucosidase family 15 | g8238 | Carbohydrate metabolic process | 52.0 | 5.2 |
| GH family 16 | g9585 | Metabolism | 36.0 | 4.9 |
| GH family 43 | g7971 | Carbohydrate metabolic process | 35.7 | 5.7 |
| GH family 25 | g6523 | Carbohydrate metabolic process | 23.8 | 5.8 |
| Cell wall components | g7617 | Structural component of the cell wall | 16.2 | 5.3 |
| g3292 | Structural component of the cell wall | 14.6 | 4.6 | |
| Lacking Signal Peptide | ||||
| GAPDH activity, NAD and NADP binding | g5740 | Oxidoreductase activity | 36.0 | 6.7 |
| ATP binding | g6856 | Gamma-actin | 41.7 | 5.7 |
| ATP binding | g10134 | ATP binding | 69.5 | 5.1 |
| ATP binding | g7817 | Hydrolase and proton-transporting ATP synthase activity | 59.5 | 8.9 |
| Peptidase | g1273 | Metallopeptidase activity | 33.8 | 5.5 |
| GH | g6997 | Enoyl-[acyl-carrier-protein] reductase (NADH) activity | 232.8 | 6.3 |
| Others | g6816 | Metal ion binding and ubiquitin-protein transferase activity | 79.4 | 7.5 |
| Others | g3420 | Phospho relay sensor kinase activity | 112.1 | 5.5 |
| Uncharacterized protein | g10276 | Uncharacterized protein | 34.3 | 7.5 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carreras, M.; Espeso, E.A.; Gutierrez-Docio, A.; Moreno-Fernandez, S.; Prodanov, M.; Hernando, M.D.; Melgarejo, P.; Larena, I. Exploring the Extracellular Macromolecular Composition of Crude Extracts of Penicillium rubens Strain 212 for Elucidation Its Mode of Action as a Biocontrol Agent. J. Fungi 2020, 6, 131. https://doi.org/10.3390/jof6030131
Carreras M, Espeso EA, Gutierrez-Docio A, Moreno-Fernandez S, Prodanov M, Hernando MD, Melgarejo P, Larena I. Exploring the Extracellular Macromolecular Composition of Crude Extracts of Penicillium rubens Strain 212 for Elucidation Its Mode of Action as a Biocontrol Agent. Journal of Fungi. 2020; 6(3):131. https://doi.org/10.3390/jof6030131
Chicago/Turabian StyleCarreras, Maria, Eduardo A. Espeso, Alba Gutierrez-Docio, Silvia Moreno-Fernandez, Marin Prodanov, Maria Dolores Hernando, Paloma Melgarejo, and Inmaculada Larena. 2020. "Exploring the Extracellular Macromolecular Composition of Crude Extracts of Penicillium rubens Strain 212 for Elucidation Its Mode of Action as a Biocontrol Agent" Journal of Fungi 6, no. 3: 131. https://doi.org/10.3390/jof6030131
APA StyleCarreras, M., Espeso, E. A., Gutierrez-Docio, A., Moreno-Fernandez, S., Prodanov, M., Hernando, M. D., Melgarejo, P., & Larena, I. (2020). Exploring the Extracellular Macromolecular Composition of Crude Extracts of Penicillium rubens Strain 212 for Elucidation Its Mode of Action as a Biocontrol Agent. Journal of Fungi, 6(3), 131. https://doi.org/10.3390/jof6030131