Chemical Constituents and Molecular Mechanism of the Yellow Phenotype of Yellow Mushroom (Floccularia luteovirens)
Abstract
:1. Introduction
2. Results
2.1. Analysis of Chemical Contents in Yellow Mushrooms
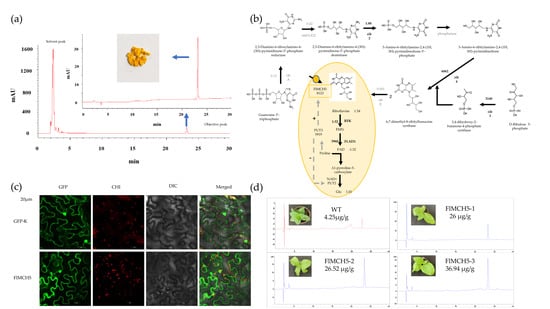
2.2. Identification and Content Determination of Riboflavin
2.3. Transcriptome Sequencing and Data Analysis
2.4. Conjoint Analysis of Transcriptome and Metabolome
2.5. Molecular Characteristics of FlMCH5
2.6. Overexpression of FlMCH5 Induces Riboflavin in Tobacco
3. Discussion
3.1. Metabolome Clarify the Main Compounds in Mushrooms
3.2. Riboflavin Is the Main Chemical Basis of the Yellow Phenotype
3.3. Riboflavin Synthesis Also Exists in White Mushroom
3.4. FlMCH5 Produces the Yellow Phenotype
3.5. FlMCH5 Is a Functional MFS Transcription Factor Gene Regulation the Yellow Phenotype
3.6. Creation of a New Disease-Resistant Tobacco with a High Riboflavin Content
4. Materials and Methods
4.1. Plant Materials
4.2. Analysis of Chemical Contents
4.3. Qualitative and Quantitative Analysis of Yellow Compound
4.4. Transcriptome Sequencing and Analysis
4.5. Functional Analysis of FlMCH5
4.6. Overexpression of FlMCH5 in Tobacco
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xing, R.; Gao, Q.B.; Zhang, F.Q.; Fu, P.C.; Wang, J.L.; Yan, H.Y.; Chen, S.L. Genetic variation and phylogenetic relationships of the ectomycorrhizal Floccularia luteovirens on the Qinghai-Tibet Plateau. J. Microbiol. 2017, 55, 600–606. [Google Scholar] [CrossRef]
- Liu, Z.; Lu, H.; Zhang, X.; Chen, Q. The Genomic and Transcriptomic Analyses of Floccularia luteovirens, a Rare Edible Fungus in the Qinghai-Tibet Plateau, Provide Insights into the Taxonomy Placement and Fruiting Body Formation. J. Fungi 2021, 7, 887. [Google Scholar] [CrossRef]
- Liu, Z.; Jiao, Y.; Lu, H.; Shu, X.; Chen, Q. Chemical characterization, antioxidant properties and anticancer activity of exopolysaccharides from Floccularia luteovirens. Carbohydr. Polym. 2020, 229, 115432. [Google Scholar] [CrossRef]
- Fu, Y.; Yu, Y.; Tan, H.; Wang, B.; Peng, W.; Sun, Q. Metabolomics reveals dopa melanin involved in the enzymatic browning of the yellow cultivars of East Asian golden needle mushroom (Flammulina filiformis). Food Chem. 2022, 370, 131295. [Google Scholar] [CrossRef]
- Ng, T.B.; Wang, H.X. A homodimeric laccase with unique characteristics from the yellow mushroom Cantharellus cibarius. Biochem Biophys Res. Commun. 2004, 313, 37–41. [Google Scholar] [CrossRef]
- Vinogradov, E.; Petersen, B.O.; Duus, J.O.; Wasser, S. The structure of the glucuronoxylomannan produced by culinary-medicinal yellow brain mushroom (Tremella mesenterica Ritz.:Fr., Heterobasidiomycetes) grown as one cell biomass in submerged culture. Carbohydr. Res. 2004, 339, 1483–1489. [Google Scholar] [CrossRef]
- Wang, H.X.; Ngai, H.K.; Ng, T.B. A ubiquitin-like peptide with ribonuclease activity against various polyhomoribonucleotides from the yellow mushroom Cantharellus cibarius. Peptides 2003, 24, 509–513. [Google Scholar] [CrossRef]
- Xing, R.; Yan, H.Y.; Gao, Q.B.; Zhang, F.Q.; Wang, J.L.; Chen, S.L. Microbial communities inhabiting the fairy ring of Floccularia luteovirens and isolation of potential mycorrhiza helper bacteria. J. Basic Microbiol. 2018, 58, 554–563. [Google Scholar] [CrossRef] [PubMed]
- Rosnina, A.G.; Tan, Y.S.; Abdullah, N.; Vikineswary, S. Morphological and molecular characterization of yellow oyster mushroom, Pleurotus citrinopileatus, hybrids obtained by interspecies mating. World J. Microbiol. Biotechnol. 2016, 32, 18. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Liu, J.; Shang, X.; Li, Y.; Zhang, L.; Li, Z.; Jiang, N.; Tan, Q.; Yu, H.; Song, C. Characterizing Diversity Based on the Chemical and Nutritional Composition of Shiitake Culinary-Medicinal Mushroom Lentinula edodes (Agaricomycetes) Commonly Cultivated in China. Int. J. Med. Mushrooms 2021, 23, 51–64. [Google Scholar] [CrossRef] [PubMed]
- Berman, J.; Sheng, Y.; Gomez Gomez, L.; Veiga, T.; Ni, X.; Farre, G.; Capell, T.; Guitian, J.; Guitian, P.; Sandmann, G.; et al. Red Anthocyanins and Yellow Carotenoids Form the Color of Orange-Flower Gentian (Gentiana lutea L. var. aurantiaca). PLoS ONE 2016, 11, e0162410. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Mizuno, T.; Shimizu, K.; Mori, S.; Fukada-Tanaka, S.; Furukawa, K.; Ishiguro, K.; Tanaka, Y.; Iida, S. Generation of Yellow Flowers of the Japanese Morning Glory by Engineering Its Flavonoid Biosynthetic Pathway toward Aurones. Plant. Cell Physiol 2019, 60, 1871–1879. [Google Scholar] [CrossRef]
- Xu, Q.; Tang, C.; Wang, X.; Sun, S.; Zhao, J.; Kang, Z.; Wang, X. An effector protein of the wheat stripe rust fungus targets chloroplasts and suppresses chloroplast function. Nat. Commun. 2019, 10, 5571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miura, E.; Kato, Y.; Matsushima, R.; Albrecht, V.; Laalami, S.; Sakamoto, W. The balance between protein synthesis and degradation in chloroplasts determines leaf variegation in Arabidopsis yellow variegated mutants. Plant. Cell 2007, 19, 1313–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suwannasom, N.; Kao, I.; Pruss, A.; Georgieva, R.; Baumler, H. Riboflavin: The Health Benefits of a Forgotten Natural Vitamin. Int. J. Mol. Sci. 2020, 21, 950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saedisomeolia, A.; Ashoori, M. Riboflavin in Human Health: A Review of Current Evidences. Adv. Food Nutr. Res. 2018, 83, 57–81. [Google Scholar] [CrossRef] [PubMed]
- Thakur, K.; Tomar, S.K.; Singh, A.K.; Mandal, S.; Arora, S. Riboflavin and health: A review of recent human research. Crit. Rev. Food Sci. Nutr. 2017, 57, 3650–3660. [Google Scholar] [CrossRef]
- Mosegaard, S.; Dipace, G.; Bross, P.; Carlsen, J.; Gregersen, N.; Olsen, R.K.J. Riboflavin Deficiency-Implications for General Human Health and Inborn Errors of Metabolism. Int. J. Mol. Sci. 2020, 21, 3847. [Google Scholar] [CrossRef]
- Lambertz, C.; Leopold, J.; Damme, K.; Vogt-Kaute, W.; Ammer, S.; Leiber, F. Effects of a riboflavin source suitable for use in organic broiler diets on performance traits and health indicators. Animal 2020, 14, 716–724. [Google Scholar] [CrossRef]
- Lehmann, M.; Degen, S.; Hohmann, H.P.; Wyss, M.; Bacher, A.; Schramek, N. Biosynthesis of riboflavin. Screening for an improved GTP cyclohydrolase II mutant. FEBS J. 2009, 276, 4119–4129. [Google Scholar] [CrossRef]
- Fassbinder, F.; Kist, M.; Bereswill, S. Structural and functional analysis of the riboflavin synthesis genes encoding GTP cyclohydrolase II (ribA), DHBP synthase (ribBA), riboflavin synthase (ribC), and riboflavin deaminase/reductase (ribD) from Helicobacter pylori strain P1. FEMS Microbiol. Lett. 2000, 191, 191–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liauta-Teglivets, O.; Hasslacher, M.; Boretskii Iu, R.; Kohlwein, S.D.; Shavlovskii, G.M. Molecular cloning of the GTP-cyclohydrolase structural gene RIB1 of Pichia guilliermondii involved in riboflavin biosynthesis. Yeast 1995, 11, 945–952. [Google Scholar] [CrossRef] [PubMed]
- Herz, S.; Eberhardt, S.; Bacher, A. Biosynthesis of riboflavin in plants. The ribA gene of Arabidopsis thaliana specifies a bifunctional GTP cyclohydrolase II/3,4-dihydroxy-2-butanone 4-phosphate synthase. Phytochemistry 2000, 53, 723–731. [Google Scholar] [CrossRef]
- Reddy, V.S.; Shlykov, M.A.; Castillo, R.; Sun, E.I.; Saier, M.H., Jr. The major facilitator superfamily (MFS) revisited. FEBS J. 2012, 279, 2022–2035. [Google Scholar] [CrossRef] [PubMed]
- Reihl, P.; Stolz, J. The monocarboxylate transporter homolog Mch5p catalyzes riboflavin (vitamin B2) uptake in Saccharomyces cerevisiae. J. Biol. Chem. 2005, 280, 39809–39817. [Google Scholar] [CrossRef] [Green Version]
- Dulyayangkul, P.; Satapoomin, N.; Avison, M.B.; Charoenlap, N.; Vattanaviboon, P.; Mongkolsuk, S. Over-Expression of Hypochlorite Inducible Major Facilitator Superfamily (MFS) Pumps Reduces Antimicrobial Drug Susceptibility by Increasing the Production of MexXY Mediated by ArmZ in Pseudomonas aeruginosa. Front. Microbiol. 2020, 11, 592153. [Google Scholar] [CrossRef]
- Pasqua, M.; Grossi, M.; Zennaro, A.; Fanelli, G.; Micheli, G.; Barras, F.; Colonna, B.; Prosseda, G. The Varied Role of Efflux Pumps of the MFS Family in the Interplay of Bacteria with Animal and Plant Cells. Microorganisms 2019, 7, 285. [Google Scholar] [CrossRef] [Green Version]
- Vishwakarma, P.; Banerjee, A.; Pasrija, R.; Prasad, R.; Lynn, A.M. Phylogenetic and conservation analyses of MFS transporters. 3 Biotech 2018, 8, 462. [Google Scholar] [CrossRef]
- Liu, J.; Fu, S.; Yang, L.; Luan, M.; Zhao, F.; Luan, S.; Lan, W. Vacuolar SPX-MFS transporters are essential for phosphate adaptation in plants. Plant Signal. Behav. 2016, 11, e1213474. [Google Scholar] [CrossRef] [Green Version]
- Obydah, W.O.; Shaker, G.A.; Samir, S.M.; El Bassiony, S.F.; Abd El Moneim, H.A. Effect of vanillic acid and exercise training on fatty liver and insulin resistance in rats: Possible role of fibroblast growth factor 21 and autophagy. Physiol. Int. 2021, 108, 412–426. [Google Scholar] [CrossRef]
- Popova, S.A.; Shevchenko, O.G.; Chukicheva, I.Y. Synthesis of new coumarin[1,3]oxazine derivatives of 7-hydroxy-6-isobornyl-4-methylcoumarin and their antioxidant activity. Chem. Biol. Drug Des. 2021, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.X.; Li, Q.; Li, H.J.; Ning, Y.M.; Zhang, R.H.; Zhang, X.J.; Li, X.L.; Xiao, W.L. Centrantheroside F, a new ionone glycoside from Centranthera grandiflora. J. Asian Nat. Prod. Res. 2021, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Umemura, K.; Ogawa, N.; Shimura, M.; Koga, J.; Usami, H.; Kono, T. Possible role of phytocassane, rice phytoalexin, in disease resistance of rice against the blast fungus Magnaporthe grisea. Biosci. Biotechnol. Biochem. 2003, 67, 899–902. [Google Scholar] [CrossRef] [Green Version]
- El-Shaheny, R.; Belal, F. Green conventional and first-order derivative fluorimetry methods for determination of trimebutine and its degradation product (eudesmic acid). Emphasis on the solvent and pH effects on their emission spectral properties. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2020, 226, 117603. [Google Scholar] [CrossRef] [PubMed]
- Vaddamanu, M.; Sathyanarayana, A.; Masaya, Y.; Sugiyama, S.; Kazuhisa, O.; Velappan, K.; Nandeshwar, M.; Hisano, K.; Tsutsumi, O.; Prabusankar, G. Acridine N-Heterocyclic Carbene Gold(I) Compounds: Tuning from Yellow to Blue Luminescence. Chem. Asian J. 2021, 16, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Wang, X.; He, Y.; Li, J.; Ma, K.; Zhang, Y.; Li, H.; Yin, C.; Zhang, Y. Stability evaluation of gardenia yellow pigment in presence of different phenolic compounds. Food Chem. 2022, 373, 131441. [Google Scholar] [CrossRef]
- Mallmann, L.P.; Tischer, B.; Vizzotto, M.; Rodrigues, E.; Manfroi, V. Comprehensive identification and quantification of unexploited phenolic compounds from red and yellow araca (Psidium cattleianum Sabine) by LC-DAD-ESI-MS/MS. Food Res. Int. 2020, 131, 108978. [Google Scholar] [CrossRef]
- Liesegang, A.; Fritsche, W.; Schlee, D.; Reinbothe, H. [Purine metabolism and riboflavin biosynthesis in microorganisms. IV. Storing of riboflavin in the cells of Candida guilliermondii (Cast.) Lang. et G]. Zeitschrift für allgemeine Mikrobiologie. 1971, 11, 121–130. [Google Scholar] [CrossRef]
- Brown, E.G.; Goodwin, T.W.; Jones, O.T. Studies on the biosynthesis of riboflavin. 4. Purine metabolism and riboflavin synthesis in Eremothecium ashbyii. Biochem. J. 1958, 68, 40–49. [Google Scholar] [CrossRef] [Green Version]
- Fedorovych, D.V.; Dmytruk, K.V.; Sibirny, A.A. Recent Advances in Construction of the Efficient Producers of Riboflavin and Flavin Nucleotides (FMN, FAD) in the Yeast Candida famata. Methods Mol. Biol. 2021, 2280, 15–30. [Google Scholar] [CrossRef]
- Jin, C.; Yao, Y.; Yonezawa, A.; Imai, S.; Yoshimatsu, H.; Otani, Y.; Omura, T.; Nakagawa, S.; Nakagawa, T.; Matsubara, K. Riboflavin Transporters RFVT/SLC52A Mediate Translocation of Riboflavin, Rather than FMN or FAD, across Plasma Membrane. Biol. Pharm. Bull. 2017, 40, 1990–1995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, X.; Cao, D.; Zhang, Z.; Cheng, S.; Wei, L.; Li, S.; Liu, B. Draft Genome Assembly of Floccularia luteovirens, an Edible and Symbiotic Mushroom on Qinghai-Tibet Plateau. G3 2020, 10, 1167–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mukai, Y.; Kamei, Y.; Liu, X.; Jiang, S.; Sugimoto, Y.; Mat Nanyan, N.S.B.; Watanabe, D.; Takagi, H. Proline metabolism regulates replicative lifespan in the yeast Saccharomyces cerevisiae. Microb. Cell 2019, 6, 482–490. [Google Scholar] [CrossRef] [PubMed]
- Nugroho, R.H.; Yoshikawa, K.; Matsuda, F.; Shimizu, H. Positive effects of proline addition on the central metabolism of wild-type and lactic acid-producing Saccharomyces cerevisiae strains. Bioprocess. Biosyst. Eng. 2016, 39, 1711–1716. [Google Scholar] [CrossRef]
- Tebung, W.A.; Omran, R.P.; Fulton, D.L.; Morschhauser, J.; Whiteway, M. Put3 Positively Regulates Proline Utilization in Candida albicans. mSphere 2017, 2, 317–354. [Google Scholar] [CrossRef] [Green Version]
- des Etages, S.A.; Falvey, D.A.; Reece, R.J.; Brandriss, M.C. Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae. Genetics 1996, 142, 1069–1082. [Google Scholar] [CrossRef]
- Spitzner, A.; Perzlmaier, A.F.; Geillinger, K.E.; Reihl, P.; Stolz, J. The proline-dependent transcription factor Put3 regulates the expression of the riboflavin transporter MCH5 in Saccharomyces cerevisiae. Genetics 2008, 180, 2007–2017. [Google Scholar] [CrossRef] [Green Version]
- Sharifi-Rad, J.; Song, S.; Ali, A.; Subbiah, V.; Taheri, Y.; Suleria, H.A.R. LC-ESI-QTOF-MS/MS characterization of phenolic compounds from Pyracantha coccinea M.Roem. and their antioxidant capacity. Cell Mol. Biol. 2021, 67, 201–211. [Google Scholar] [CrossRef]
- Puffeld, M.; Seiler, C.; Kuhlmann, M.; Sreenivasulu, N.; Butardo, V.M., Jr. Analysis of Developing Rice Grain Transcriptome Using the Agilent Microarray Platform. Methods Mol. Biol. 2019, 1892, 277–300. [Google Scholar] [CrossRef]
- Domingo-Almenara, X.; Siuzdak, G. Metabolomics Data Processing Using XCMS. Methods Mol. Biol. 2020, 2104, 11–24. [Google Scholar] [CrossRef]
- Saccenti, E.; Timmerman, M.E. Approaches to Sample Size Determination for Multivariate Data: Applications to PCA and PLS-DA of Omics Data. J. Proteome Res. 2016, 15, 2379–2393. [Google Scholar] [CrossRef]
- Worley, B.; Halouska, S.; Powers, R. Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal. Biochem. 2013, 433, 102–104. [Google Scholar] [CrossRef] [Green Version]
- Khandakar, A.; Chowdhury, M.E.H.; Ibne Reaz, M.B.; Md Ali, S.H.; Hasan, M.A.; Kiranyaz, S.; Rahman, T.; Alfkey, R.; Bakar, A.A.A.; Malik, R.A. A machine learning model for early detection of diabetic foot using thermogram images. Comput. Biol. Med. 2021, 137, 104838. [Google Scholar] [CrossRef] [PubMed]
- Schwechheimer, S.K.; Becker, J.; Peyriga, L.; Portais, J.C.; Sauer, D.; Muller, R.; Hoff, B.; Haefner, S.; Schroder, H.; Zelder, O.; et al. Improved riboflavin production with Ashbya gossypii from vegetable oil based on (13)C metabolic network analysis with combined labeling analysis by GC/MS, LC/MS, 1D, and 2D NMR. Metab. Eng. 2018, 47, 357–373. [Google Scholar] [CrossRef] [PubMed]
- Deng, K.; Ouyang, J.; Hu, N.; Meng, J.; Su, C.; Wang, J.; Wang, H. Improved colorimetric analysis for subtle changes of powdered anthocyanins extracted from Lycium ruthenicum Murr. Food Chem. 2022, 371, 131080. [Google Scholar] [CrossRef] [PubMed]
- Villa-Rodriguez, E.; Ibarra-Gamez, C.; de Los Santos-Villalobos, S. Extraction of high-quality RNA from Bacillus subtilis with a lysozyme pre-treatment followed by the Trizol method. J. Microbiol. Methods 2018, 147, 14–16. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef]
- Kim, C.S.; Winn, M.D.; Sachdeva, V.; Jordan, K.E. K-mer clustering algorithm using a MapReduce framework: Application to the parallelization of the Inchworm module of Trinity. BMC Bioinform. 2017, 18, 467. [Google Scholar] [CrossRef] [Green Version]
- Yu, D.S.; Lee, D.H.; Kim, S.K.; Lee, C.H.; Song, J.Y.; Kong, E.B.; Kim, J.F. Algorithm for predicting functionally equivalent proteins from BLAST and HMMER searches. J. Microbiol. Biotechnol. 2012, 22, 1054–1058. [Google Scholar] [CrossRef]
- Zhu, D.; Hero, A.O.; Qin, Z.S.; Swaroop, A. High throughput screening of co-expressed gene pairs with controlled false discovery rate (FDR) and minimum acceptable strength (MAS). J. Comput. Biol. 2005, 12, 1029–1045. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zong, Y.; Xi, X.; Li, S.; Chen, W.; Zhang, B.; Liu, D.; Liu, B.; Wang, D.; Zhang, H. Allelic Variation and Transcriptional Isoforms of Wheat TaMYC1 Gene Regulating Anthocyanin Synthesis in Pericarp. Front. Plant. Sci 2017, 8, 1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, G.; Moriyama, E.N. Vector NTI, a balanced all-in-one sequence analysis suite. Brief. Bioinform. 2004, 5, 378–388. [Google Scholar] [CrossRef]
- Zong, Y.; Li, S.; Xi, X.; Cao, D.; Wang, Z.; Wang, R.; Liu, B. Comprehensive Influences of Overexpression of a MYB Transcriptor Regulating Anthocyanin Biosynthesis on Transcriptome and Metabolome of Tobacco Leaves. Int. J. Mol. Sci. 2019, 20, 5123. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Class | Total | Down (in Yellow) | Up (in Yellow) | Number Different | Ratio (Yellow/White) |
|---|---|---|---|---|---|
| Amino acid and derivatives | 98 | 22 | 23 | 45 | 0.466 |
| Phenylpropanoids | 35 | 13 | 7 | 20 | 0.278 |
| Flavone | 35 | 1 | 8 | 9 | 8.758 |
| Flavonol | 16 | 2 | 1 | 3 | 3.691 |
| Flavonoid | 16 | 1 | 1 | 2 | 0.339 |
| Flavanone | 11 | 2 | 1 | 3 | 1.392 |
| Isoflavone | 2 | 1 | 1 | 2 | 2.149 |
| Alcohols | 15 | 5 | 0 | 5 | 0.005 |
| Polyphenol | 12 | 1 | 1 | 2 | 0.046 |
| Phenolamides | 12 | 2 | 4 | 6 | 1.969 |
| Nucleotide and derivates | 59 | 20 | 13 | 33 | 1.167 |
| Others | 27 | 6 | 6 | 12 | 0.387 |
| Alkaloids | 28 | 8 | 0 | 8 | 0.103 |
| Carbohydrates | 22 | 2 | 8 | 10 | 2.446 |
| Terpene | 11 | 0 | 3 | 3 | 2.425 |
| Vitamins and derivatives | 17 | 1 | 6 | 7 | 3.362 |
| Indole derivatives | 7 | 3 | 0 | 3 | 0.315 |
| Organic acids and derivatives | 81 | 12 | 11 | 23 | 1.132 |
| Lipids | 61 | 12 | 18 | 30 | 2.274 |
| Anthocyanins | 2 | 1 | 0 | 1 | 0 |
| Quinones | 1 | 0 | 0 | 0 | / |
| Sterides | 5 | 0 | 0 | 0 | / |
| sum | 573 | 115 | 112 | 227 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, X.; Bao, X.; Liu, B.; Li, Y.; Cao, D.; Zhang, H.; Zong, Y. Chemical Constituents and Molecular Mechanism of the Yellow Phenotype of Yellow Mushroom (Floccularia luteovirens). J. Fungi 2022, 8, 314. https://doi.org/10.3390/jof8030314
Gan X, Bao X, Liu B, Li Y, Cao D, Zhang H, Zong Y. Chemical Constituents and Molecular Mechanism of the Yellow Phenotype of Yellow Mushroom (Floccularia luteovirens). Journal of Fungi. 2022; 8(3):314. https://doi.org/10.3390/jof8030314
Chicago/Turabian StyleGan, Xiaolong, Xuemei Bao, Baolong Liu, Yun Li, Dong Cao, Hg Zhang, and Yuan Zong. 2022. "Chemical Constituents and Molecular Mechanism of the Yellow Phenotype of Yellow Mushroom (Floccularia luteovirens)" Journal of Fungi 8, no. 3: 314. https://doi.org/10.3390/jof8030314
APA StyleGan, X., Bao, X., Liu, B., Li, Y., Cao, D., Zhang, H., & Zong, Y. (2022). Chemical Constituents and Molecular Mechanism of the Yellow Phenotype of Yellow Mushroom (Floccularia luteovirens). Journal of Fungi, 8(3), 314. https://doi.org/10.3390/jof8030314