From Saccharomyces cerevisiae to Ethanol: Unlocking the Power of Evolutionary Engineering in Metabolic Engineering Applications
Abstract
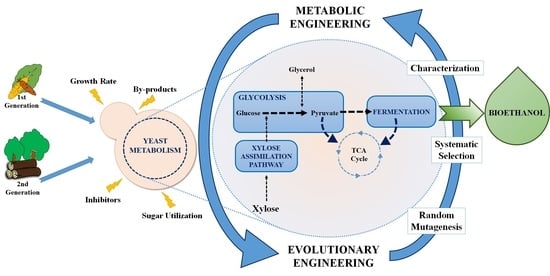
:1. Introduction
2. Factors Affecting Yeast Metabolism in Bioethanol Production
3. Metabolic Engineering of Yeast for Bioethanol Production
3.1. Lowering ATP Yield
3.2. Sustainable Reduction of Glycerol Formation
3.3. Prevention of Bacterial Contamination
3.4. Introduction and Optimization of Xylose Assimilation Pathway
3.5. Increasing Stress Tolerance
4. Evolutionary Engineering of Yeast for Bioethanol Production
4.1. Increasing Growth Rate and Viability
4.2. Decreasing By-Product Formation
4.3. Improving Utilization and Transport of Sugars
4.4. Increasing Tolerance to Ethanol and Lignocellulosic Inhibitors
5. Challenges of Evolutionary Engineering for Bioethanol Production
6. Future Directions for Evolution-Based Metabolic Engineering of Yeast for Bioethanol Production
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Devi, A.; Bajar, S.; Sihag, P.; Sheikh, Z.U.; Singh, A.; Kaur, J.; Bishnoi, N.R.; Pant, D. A panoramic view of technological landscape for bioethanol production from various generations of feedstocks. Bioengineered 2023, 14, 81–112. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chang, X. Past, present, and future perspectives on whey as a promising feedstock for bioethanol production by yeast. J. Fungi 2022, 8, 395. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, E.; Dussap, C.G. First Generation Bioethanol: Fundamentals—Definition, History, Global Production, Evolution. In Liquid Biofuels: Bioethanol; Soccol, C.R., Amarante Guimarães Pereira, G., Dussap, C.G., Porto de Souza Vandenberghe, L., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 1–12. [Google Scholar]
- Bušić, A.; Marđetko, N.; Kundas, S.; Morzak, G.; Belskaya, H.; Ivančić Šantek, M.; Komes, D.; Novak, S.; Šantek, B. Bioethanol production from renewable raw materials and its separation and purification: A review. Food Technol. Biotechnol. 2018, 56, 289–311. [Google Scholar] [CrossRef]
- Deng, W.; Feng, Y.; Fu, J.; Guo, H.; Guo, Y.; Han, B.; Jiang, Z.; Kong, L.; Li, C.; Liu, H.; et al. Catalytic conversion of lignocellulosic biomass into chemicals and fuels. Green Energy Environ. 2022, 8, 10–114. [Google Scholar] [CrossRef]
- Nijland, J.G.; Driessen, A.J. Engineering of pentose transport in Saccharomyces cerevisiae for biotechnological applications. Front. Bioeng. Biotechnol. 2020, 7, 464. [Google Scholar] [CrossRef] [PubMed]
- Çakar, Z.P.; Turanlı-Yıldız, B.; Alkım, C.; Yılmaz, Ü. Evolutionary engineering of Saccharomyces cerevisiae for improved industrially important properties. FEMS Yeast Res. 2012, 12, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Zabed, H.; Faruq, G.; Sahu, J.N.; Azirun, M.S.; Hashim, R.; Nasrulhaq Boyce, A. Bioethanol production from fermentable sugar juice. Sci. World J. 2014, 12, 2014. [Google Scholar] [CrossRef] [PubMed]
- Della-Bianca, B.E.; Basso, T.O.; Stambuk, B.U.; Basso, L.C.; Gombert, A.K. What do we know about the yeast strains from the Brazilian fuel ethanol industry? Appl. Microbiol. Biotechnol. 2013, 97, 979–991. [Google Scholar] [CrossRef]
- Ruchala, J.; Kurylenko, O.O.; Dmytruk, K.V.; Sibirny, A.A. Construction of advanced producers of first-and second-generation ethanol in Saccharomyces cerevisiae and selected species of non-conventional yeasts (Scheffersomyces stipitis, Ogataea polymorpha). J. Ind. Microbiol. Biotechnol. 2020, 47, 109–132. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.E. Toward a science of metabolic engineering. Science 1991, 252, 1668–1675. [Google Scholar] [CrossRef] [PubMed]
- Bailey, J.E.; Sburlati, A.; Hatzimanikatis, V.; Lee, K.; Renner, W.A.; Tsai, P.S. Inverse metabolic engineering: A strategy for directed genetic engineering of useful phenotypes. Biotechnol. Bioeng. 1996, 52, 109–121. [Google Scholar] [CrossRef]
- Butler, P.R.; Brown, M.; Oliver, S.G. Improvement of antibiotic titers from Streptomyces bacteria by interactive continuous selection. Biotechnol. Bioeng. 1996, 49, 185–196. [Google Scholar] [CrossRef]
- Sauer, U. Evolutionary engineering of industrially important microbial phenotypes. Adv. Biochem. Eng. Biotechnol. 2001, 73, 130–166. [Google Scholar]
- Sonderegger, M.; Sauer, U. Evolutionary engineering of Saccharomyces cerevisiae for anaerobic growth on xylose. Appl. Environ. Microbiol. 2003, 69, 1990–1998. [Google Scholar] [CrossRef] [PubMed]
- Guimaraes, P.M.R.; François, J.; Parrou, J.L.; Teixeira, J.A.; Domingues, L. Adaptive evolution of a lactose-consuming Saccharomyces cerevisiae recombinant. Appl. Environ. Microbiol. 2008, 74, 1748–1756. [Google Scholar] [CrossRef] [PubMed]
- Basso, T.O.; de Kok, S.; Dario, M.; do Espirito-Santo, J.C.; Müller, G.; Schlölg, P.S.; Silva, C.P.; Tonso, A.; Daran, J.M.; Gombert, A.K.; et al. Engineering topology and kinetics of sucrose metabolism in Saccharomyces cerevisiae for improved ethanol yield. Metab. Eng. 2011, 13, 694–703. [Google Scholar] [CrossRef]
- Gancedo, C.; Serrano, R. Energy-Yielding Metabolism. In The Yeasts, 2nd ed.; Rose, A.H., Harrison, J.S., Eds.; Academic Press: London, UK, 1989; Volume 3, pp. 205–259. [Google Scholar]
- Raj, S.B.; Ramaswamy, S.; Plapp, B.V. Yeast alcohol dehydrogenase structure and catalysis. Biochemistry 2014, 53, 5791–5803. [Google Scholar] [CrossRef]
- Kwak, S.; Jin, Y.S. Production of fuels and chemicals from xylose by engineered Saccharomyces cerevisiae: A review and perspective. Microb. Cell Factories 2017, 16, 82. [Google Scholar] [CrossRef]
- Cot, M.; Loret, M.O.; François, J.; Benbadis, L. Physiological behaviour of Saccharomyces cerevisiae in aerated fed-batch fermentation for high level production of bioethanol. FEMS Yeast Res. 2007, 7, 22–32. [Google Scholar] [CrossRef]
- Liu, R.; Shen, F. Impacts of main factors on bioethanol fermentation from stalk juice of sweet sorghum by immobilized Saccharomyces cerevisiae (CICC 1308). Bioresour. Technol. 2008, 99, 847–854. [Google Scholar] [CrossRef]
- Bayrock, D.P.; Michael Ingledew, W. Application of multistage continuous fermentation for production of fuel alcohol by very-high-gravity fermentation technology. J. Ind. Microbiol. Biotechnol. 2001, 27, 87–93. [Google Scholar] [CrossRef]
- Laopaiboon, L.; Nuanpeng, S.; Srinophakun, P.; Klanrit, P.; Laopaiboon, P. Ethanol production from sweet sorghum juice using very high gravity technology: Effects of carbon and nitrogen supplementations. Bioresour. Technol. 2009, 100, 4176–4182. [Google Scholar] [CrossRef] [PubMed]
- Casey, G.P.; Ingledew, W.M. Ethanol tolerance in yeasts. Crit. Rev. Microbiol. 1986, 13, 219–280. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Zhang, W.; Li, C.; Sakakibara, K.; Tanaka, S.; Kong, H. Factors affecting ethanol fermentation using Saccharomyces cerevisiae BY4742. Biomass Bioenergy 2012, 47, 395–401. [Google Scholar] [CrossRef]
- Casalta, E.; Aguera, E.; Picou, C.; Rodriguez-Bencomo, J.J.; Salmon, J.M.; Sablayrolles, J.M. A comparison of laboratory and pilot-scale fermentations in winemaking conditions. Appl. Microbiol. Biotechnol. 2010, 87, 1665–1673. [Google Scholar] [CrossRef] [PubMed]
- Crépin, L.; Sanchez, I.; Nidelet, T.; Dequin, S.; Camarasa, C. Efficient ammonium uptake and mobilization of vacuolar arginine by Saccharomyces cerevisiae wine strains during wine fermentation. Microb. Cell Factories 2014, 13, 109. [Google Scholar] [CrossRef]
- Blateyron, L.; Sablayrolles, J.M. Stuck and slow fermentations in enology: Statistical study of causes and effectiveness of combined additions of oxygen and diammonium phosphate. J. Biosci. Bioeng. 2001, 91, 184–189. [Google Scholar] [CrossRef]
- Julien, A.; Roustan, J.L.; Dulau, L.; Sablayrolles, J.M. Comparison of nitrogen and oxygen demands of enological yeasts: Technological consequences. Am. J. Enol. Vitic. 2000, 51, 215–222. [Google Scholar] [CrossRef]
- Rollero, S.; Roberts, S.; Bauer, F.F.; Divol, B. Agitation impacts fermentation performance as well as carbon and nitrogen metabolism in Saccharomyces cerevisiae under winemaking conditions. Aust. J. Grape Wine Res. 2018, 24, 360–367. [Google Scholar] [CrossRef]
- Hashem, M.; Alamri, S.A.; Asseri, T.A.; Mostafa, Y.S.; Lyberatos, G.; Ntaikou, I. On the optimization of fermentation conditions for enhanced bioethanol yields from starchy biowaste via yeast co-cultures. Sustainability 2021, 13, 1890. [Google Scholar] [CrossRef]
- Laopaiboon, L.; Thanonkeo, P.; Jaisil, P.; Laopaiboon, P. Ethanol production from sweet sorghum juice in batch and fed-batch fermentations by Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2007, 23, 1497–1501. [Google Scholar] [CrossRef]
- Breisha, G.Z. Production of 16% ethanol from 35% sucrose. Biomass Bioenergy 2010, 34, 1243–1249. [Google Scholar] [CrossRef]
- Klinke, H.B.; Thomsen, A.B.; Ahring, B.K. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl. Microbiol. Biotechnol. 2004, 66, 10–26. [Google Scholar] [CrossRef]
- Chen, H.; Liu, J.; Chang, X.; Chen, D.; Xue, Y.; Liu, P.; Lin, H.; Han, S. A review on the pretreatment of lignocellulose for high-value chemicals. Fuel Process. Technol. 2017, 160, 196–206. [Google Scholar] [CrossRef]
- Kim, S.K.; Jin, Y.S.; Choi, I.G.; Park, Y.C.; Seo, J.H. Enhanced tolerance of Saccharomyces cerevisiae to multiple lignocellulose-derived inhibitors through modulation of spermidine contents. Metab. Eng. 2015, 29, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, E.C.; Bakker, R.R.; Baets, P.; Eggink, G. By-products resulting from lignocellulose pretreatment and their inhibitory effect on fermentations for (bio) chemicals and fuels. Appl. Microbiol. Biotechnol. 2014, 98, 9579–9593. [Google Scholar] [CrossRef] [PubMed]
- Fraczek, M.G.; Naseeb, S.; Delneri, D. History of genome editing in yeast. Yeast 2018, 35, 361–368. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.A.; Vasser, M.; Powers, D.B. Cassette mutagenesis: An efficient method for generation of multiple mutations at defined sites. Gene 1985, 34, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Ingledew, W.M. Alcohol production by Saccharomyces cerevisiae: A yeast primer. In The Alcohol Textbook: A Reference for the Beverage, Fuel and Industrial Alcohol Industries; 3rd ed., Jacques, K.A., Lyons, T.P., Kelsall, D.R., Eds.; Nottingham University Press: Nottingham, UK, 1999; Volume 3, pp. 49–87. [Google Scholar]
- Van Aalst, A.C.; de Valk, S.C.; van Gulik, W.M.; Jansen, M.L.; Pronk, J.T.; Mans, R. Pathway engineering strategies for improved product yield in yeast-based industrial ethanol production. Synth. Syst. Biotechnol. 2022, 7, 554–566. [Google Scholar] [CrossRef]
- Katz, J.; Rognstad, R. Futile cycling in glucose metabolism. Trends Biochem. Sci. 1978, 3, 171–174. [Google Scholar] [CrossRef]
- Semkiv, M.V.; Dmytruk, K.V.; Abbas, C.A.; Sibirny, A.A. Activation of futile cycles as an approach to increase ethanol yield during glucose fermentation in Saccharomyces cerevisiae. Bioengineered 2016, 7, 106–111. [Google Scholar] [CrossRef]
- Zahoor, A.; Messerschmidt, K.; Boecker, S.; Klamt, S. ATPase-based implementation of enforced ATP wasting in Saccharomyces cerevisiae for improved ethanol production. Biotechnol. Biofuels 2020, 13, 185. [Google Scholar] [CrossRef] [PubMed]
- Semkiv, M.V.; Dmytruk, K.V.; Abbas, C.A.; Sibirny, A.A. Increased ethanol accumulation from glucose via reduction of ATP level in a recombinant strain of Saccharomyces cerevisiae overexpressing alkaline phosphatase. BMC Biotechnol. 2014, 14, 42. [Google Scholar] [CrossRef] [PubMed]
- Sprenger, G.A. Carbohydrate metabolism in Zymomonas mobilis: A catabolic highway with some scenic routes. FEMS Microbiol. Lett. 1996, 145, 301–307. [Google Scholar] [CrossRef]
- Benisch, F.; Boles, E. The bacterial Entner–Doudoroff pathway does not replace glycolysis in Saccharomyces cerevisiae due to the lack of activity of iron–sulfur cluster enzyme 6-phosphogluconate dehydratase. J. Biotechnol. 2014, 171, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Morita, K.; Nomura, Y.; Ishii, J.; Matsuda, F.; Kondo, A.; Shimizu, H. Heterologous expression of bacterial phosphoenol pyruvate carboxylase and Entner–Doudoroff pathway in Saccharomyces cerevisiae for improvement of isobutanol production. J. Biosci. Bioeng. 2017, 124, 263–270. [Google Scholar] [CrossRef]
- Gombert, A.K.; van Maris, A.J. Improving conversion yield of fermentable sugars into fuel ethanol in 1st generation yeast-based production processes. Curr. Opin. Biotechnol. 2015, 33, 81–86. [Google Scholar] [CrossRef]
- Nissen, T.L.; Hamann, C.W.; Kielland-Brandt, M.C.; Nielsen, J.; Villadsen, J. Anaerobic and aerobic batch cultivations of Saccharomyces cerevisiae mutants impaired in glycerol synthesis. Yeast 2000, 16, 463–474. [Google Scholar] [CrossRef]
- Van Dijken, J.P.; Scheffers, W.A. Redox balances in the metabolism of sugars by yeasts. FEMS Microbiol. Rev. 1986, 1, 199–224. [Google Scholar] [CrossRef]
- Eriksson, P.; André, L.; Ansell, R.; Blomberg, A.; Adler, L. Cloning and characterization of GPD2, a second gene encoding sn-glycerol 3-phosphate dehydrogenase (NAD+) in Saccharomyces cerevisiae, and its comparison with GPD1. Mol. Microbiol. 1995, 17, 95–107. [Google Scholar] [CrossRef]
- Hubmann, G.; Guillouet, S.; Nevoigt, E. Gpd1 and Gpd2 fine-tuning for sustainable reduction of glycerol formation in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2011, 77, 5857–5867. [Google Scholar] [CrossRef] [PubMed]
- Pagliardini, J.; Hubmann, G.; Alfenore, S.; Nevoigt, E.; Bideaux, C.; Guillouet, S.E. The metabolic costs of improving ethanol yield by reducing glycerol formation capacity under anaerobic conditions in Saccharomyces cerevisiae. Microb. Cell Factories 2013, 12, 29. [Google Scholar] [CrossRef]
- Jain, V.K.; Divol, B.; Prior, B.A.; Bauer, F.F. Elimination of glycerol and replacement with alternative products in ethanol fermentation by Saccharomyces cerevisiae. J. Ind. Microbiol. Biotechnol. 2011, 38, 1427–1435. [Google Scholar] [CrossRef]
- Zhang, L.; Tang, Y.; Guo, Z.P.; Ding, Z.Y.; Shi, G.Y. Improving the ethanol yield by reducing glycerol formation using cofactor regulation in Saccharomyces cerevisiae. Biotechnol. Lett. 2011, 33, 1375–1380. [Google Scholar] [CrossRef] [PubMed]
- Nissen, T.L.; Kielland-Brandt, M.C.; Nielsen, J.; Villadsen, J. Optimization of ethanol production in Saccharomyces cerevisiae by metabolic engineering of the ammonium assimilation. Metab. Eng. 2000, 2, 69–77. [Google Scholar] [CrossRef]
- Papapetridis, I.; Goudriaan, M.; Vázquez Vitali, M.; De Keijzer, N.A.; Van Den Broek, M.; Van Maris, A.J.; Pronk, J.T. Optimizing anaerobic growth rate and fermentation kinetics in Saccharomyces cerevisiae strains expressing Calvin-cycle enzymes for improved ethanol yield. Biotechnol. Biofuels 2018, 11, 17. [Google Scholar] [CrossRef] [PubMed]
- Hohmann, S. Osmotic stress signaling and osmoadaptation in yeasts. Microbiol. Mol. Biol. Rev. 2002, 66, 300–372. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.P.; Zhang, L.; Ding, Z.Y.; Shi, G.Y. Minimization of glycerol synthesis in industrial ethanol yeast without influencing its fermentation performance. Metab. Eng. 2011, 13, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.A.; Hirasawa, T.; Shimizu, H. Differential importance of trehalose accumulation in Saccharomyces cerevisiae in response to various environmental stresses. J. Biosci. Bioeng. 2010, 109, 262–266. [Google Scholar] [CrossRef] [PubMed]
- An, M.Z.; Tang, Y.Q.; Mitsumasu, K.; Liu, Z.S.; Shigeru, M.; Kenji, K. Enhanced thermotolerance for ethanol fermentation of Saccharomyces cerevisiae strain by overexpression of the gene coding for trehalose-6-phosphate synthase. Biotechnol. Lett. 2011, 33, 1367–1374. [Google Scholar] [CrossRef] [PubMed]
- Shahsavarani, H.; Sugiyama, M.; Kaneko, Y.; Chuenchit, B.; Harashima, S. Superior thermotolerance of Saccharomyces cerevisiae for efficient bioethanol fermentation can be achieved by overexpression of RSP5 ubiquitin ligase. Biotechnol. Adv. 2012, 30, 1289–1300. [Google Scholar] [CrossRef] [PubMed]
- Swinnen, S.; Schaerlaekens, K.; Pais, T.; Claesen, J.; Hubmann, G.; Yang, Y.; Demeke, M.; Foulquié-Moreno, M.R.; Goovaerts, A.; Souvereyns, K.; et al. Identification of novel causative genes determining the complex trait of high ethanol tolerance in yeast using pooled-segregant whole-genome sequence analysis. Genome Res. 2012, 22, 975–984. [Google Scholar] [CrossRef] [PubMed]
- Varize, C.S.; Bücker, A.; Lopes, L.D.; Christofoleti-Furlan, R.M.; Raposo, M.S.; Basso, L.C.; Stambuk, B.U. Increasing ethanol tolerance and ethanol production in an industrial fuel ethanol Saccharomyces cerevisiae strain. Fermentation 2022, 8, 470. [Google Scholar] [CrossRef]
- Basso, L.C.; de Amorim, H.V.; de Oliveira, A.J.; Lopes, M.L. Yeast selection for fuel ethanol production in Brazil. FEMS Yeast Res. 2008, 8, 1155–1163. [Google Scholar] [CrossRef] [PubMed]
- Bischoff, K.M.; Skinner-Nemec, K.A.; Leathers, T.D. Antimicrobial susceptibility of Lactobacillus species isolated from commercial ethanol plants. J. Ind. Microbiol. Biotechnol. 2007, 34, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Beckner, M.; Ivey, M.L.; Phister, T.G. Microbial contamination of fuel ethanol fermentations. Lett. Appl. Microbiol. 2011, 53, 387–394. [Google Scholar] [CrossRef]
- Jansen, M.L.A.; Bracher, J.M.; Papapetridis, I.; Verhoeven, M.D.; de Bruijn, H.; de Waal, P.P.; van Maris, A.J.A.; Klaassen, P.; Pronk, J.T. Saccharomyces cerevisiae strains for second-generation ethanol production: From academic exploration to industrial implementation. FEMS Yeast Res. 2017, 17, fox044. [Google Scholar] [CrossRef]
- Inaba, T.; Watanabe, D.; Yoshiyama, Y.; Tanaka, K.; Ogawa, J.; Takagi, H.; Shimoi, H.; Shima, J. An organic acid-tolerant HAA1-overexpression mutant of an industrial bioethanol strain of Saccharomyces cerevisiae and its application to the production of bioethanol from sugarcane molasses. AMB Express. 2013, 3, 74. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.Y.; Liu, S.; Patel, M.H.; Glenzinski, K.M.; Skory, C.D. Saccharomyces cerevisiae surface display of endolysin LysKB317 for control of bacterial contamination in corn ethanol fermentations. Front. Bioeng. Biotechnol. 2023, 11, 1162720. [Google Scholar] [CrossRef]
- Kim, S.R.; Park, Y.C.; Jin, Y.S.; Seo, J.H. Strain engineering of Saccharomyces cerevisiae for enhanced xylose metabolism. Biotechnol. Adv. 2013, 31, 851–861. [Google Scholar] [CrossRef]
- Eliasson, A.; Christensson, C.; Wahlbom, C.F.; Hahn-Hägerdal, B. Anaerobic xylose fermentation by recombinant Saccharomyces cerevisiae carrying XYL1, XYL2, and XKS1 in mineral medium chemostat cultures. Appl. Environ. Microbiol. 2000, 66, 3381–3386. [Google Scholar] [CrossRef]
- Harhangi, H.R.; Akhmanova, A.S.; Emmens, R.; van der Drift, C.; de Laat, W.T.; van Dijken, J.P.; Jetten, M.S.; Pronk, J.T.; Op den Camp, H.J. Xylose metabolism in the anaerobic fungus Piromyces sp. strain E2 follows the bacterial pathway. Arch. Microbiol. 2003, 180, 134–141. [Google Scholar] [CrossRef]
- Lee, S.M.; Jellison, T.; Alper, H.S. Directed evolution of xylose isomerase for improved xylose catabolism and fermentation in the yeast Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 5708–5716. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, A.; Van Zyl, C.; Van Tonder, A.; Prior, B.A. Purification and partial characterization of an aldo-keto reductase from Saccharomyces cerevisiae. Appl. Environ. Microbiol. 1995, 61, 1580–1585. [Google Scholar] [CrossRef]
- Toivari, M.H.; Salusja, L.; Ruohonen, L.; Penttila, M. Endogenous xylose pathway in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2004, 70, 3681–3686. [Google Scholar] [CrossRef]
- Romaní, A.; Pereira, F.; Johansson, B.; Domingues, L. Metabolic engineering of Saccharomyces cerevisiae ethanol strains PE-2 and CAT-1 for efficient lignocellulosic fermentation. Bioresour. Technol. 2015, 179, 150–158. [Google Scholar] [CrossRef]
- Li, Y.C.; Mitsumasu, K.; Gou, Z.X.; Gou, M.; Tang, Y.Q.; Li, G.Y.; Wu, X.L.; Akamatsu, T.; Taguchi, H.; Kida, K. Xylose fermentation efficiency and inhibitor tolerance of the recombinant industrial Saccharomyces cerevisiae strain NAPX37. Appl. Microbiol. Biotechnol. 2016, 100, 1531–1542. [Google Scholar] [CrossRef]
- Cadete, R.M.; de Las Heras, A.M.; Sandström, A.G.; Ferreira, C.; Gírio, F.; Gorwa-Grauslund, M.F.; Rosa, C.A.; Fonseca, C. Exploring xylose metabolism in Spathaspora species: XYL1. 2 from Spathaspora passalidarum as the key for efficient anaerobic xylose fermentation in metabolic engineered Saccharomyces cerevisiae. Biotechnol. Biofuels. 2016, 9, 167. [Google Scholar] [CrossRef] [PubMed]
- Xia, P.F.; Zhang, G.C.; Walker, B.; Seo, S.O.; Kwak, S.; Liu, J.J.; Kim, H.; Ort, D.R.; Wang, S.G.; Jin, Y.S. Recycling carbon dioxide during xylose fermentation by engineered Saccharomyces cerevisiae. ACS Synth. Biol. 2017, 6, 276–283. [Google Scholar] [CrossRef] [PubMed]
- De Figueiredo Vilela, L.; de Mello, V.M.; Reis, V.C.; da Silva Bon, E.P.; Torres, F.A.; Neves, B.C.; Eleutherio, E.C. Functional expression of Burkholderia cenocepacia xylose isomerase in yeast increases ethanol production from a glucose–xylose blend. Bioresour. Technol. 2013, 128, 792–796. [Google Scholar] [CrossRef]
- Li, Y.C.; Li, G.Y.; Gou, M.; Xia, Z.Y.; Tang, Y.Q.; Kida, K. Functional expression of xylose isomerase in flocculating industrial Saccharomyces cerevisiae strain for bioethanol production. J. Biosci. Bioeng. 2016, 121, 685–691. [Google Scholar] [CrossRef] [PubMed]
- Temer, B.; Dos Santos, L.V.; Negri, A.; Galhardo, J.P.; Magalhães, P.H.; José, J.; Marschalk, C.; Corrêa, T.L.; Carazzolle, M.F.; Pereira, G.A. Conversion of an inactive xylose isomerase into a functional enzyme by co-expression of GroEL-GroES chaperonins in Saccharomyces cerevisiae. BMC Biotechnol. 2017, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- De Souza Colombo, G.; Viana Mendes, I.; de Morais Souto, B.; Chaves Barreto, C.; Assis Serra, L.; Ferreira Noronha, E.; Skorupa Parachin, N.; Moreira de Almeida, J.R.; Ferraz Quirino, B. Identification and functional expression of a new xylose isomerase from the goat rumen microbiome in Saccharomyces cerevisiae. Lett. Appl. Microbiol. 2022, 74, 941–948. [Google Scholar] [CrossRef]
- Favaro, L.; Jansen, T.; van Zyl, W.H. Exploring industrial and natural Saccharomyces cerevisiae strains for the bio-based economy from biomass: The case of bioethanol. Crit. Rev. Biotechnol. 2019, 3, 800–816. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Wu, Y.; Zheng, Z.; Cao, L.; Zhu, X.; Mu, D.; Jiang, S. CRISPR-Cas9 approach constructing cellulase sestc-engineered Saccharomyces cerevisiae for the production of orange peel ethanol. Front. Microbiol. 2018, 9, 2436. [Google Scholar] [CrossRef] [PubMed]
- Arnthong, J.; Ponjarat, J.; Bussadee, P.; Deenarn, P.; Prommana, P.; Phienluphon, A.; Charoensri, S.; Champreda, V.; Zhao, X.Q.; Suwannarangsee, S. Enhanced surface display efficiency of β-glucosidase in Saccharomyces cerevisiae by disruption of cell wall protein-encoding genes YGP1 and CWP2. Biochem. Eng. J. 2022, 179, 108305. [Google Scholar] [CrossRef]
- Sjulander, N.; Kikas, T. Origin, impact and control of lignocellulosic inhibitors in bioethanol production—A review. Energies 2020, 13, 4751. [Google Scholar] [CrossRef]
- Almeida, J.R.; Röder, A.; Modig, T.; Laadan, B.; Lidén, G.; Gorwa-Grauslund, M.F. NADH-vs NADPH-coupled reduction of 5-hydroxymethyl furfural (HMF) and its implications on product distribution in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2008, 78, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Vanmarcke, G.; Deparis, Q.; Vanthienen, W.; Peetermans, A.; Foulquié-Moreno, M.R.; Thevelein, J.M. A novel AST2 mutation generated upon whole-genome transformation of Saccharomyces cerevisiae confers high tolerance to 5-Hydroxymethylfurfural (HMF) and other inhibitors. PLoS Genet. 2021, 17, e1009826. [Google Scholar] [CrossRef] [PubMed]
- Gorsich, S.W.; Dien, B.S.; Nichols, N.N.; Slininger, P.J.; Liu, Z.L.; Skory, C.D. Tolerance to furfural-induced stress is associated with pentose phosphate pathway genes ZWF1, GND1, RPE1, and TKL1 in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2006, 71, 339–349. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, K.; Mehmood, M.A.; Zhao, Z.K.; Bai, F.; Zhao, X. Deletion of acetate transporter gene ADY2 improved tolerance of Saccharomyces cerevisiae against multiple stresses and enhanced ethanol production in the presence of acetic acid. Bioresour. Technol. 2017, 245, 1461–1468. [Google Scholar] [CrossRef]
- Milessi, T.S.; Perez, C.L.; Zangirolami, T.C.; Corradini, F.A.; Sandri, J.P.; Foulquié-Moreno, M.R.; Giordano, R.C.; Thevelein, J.M.; Giordano, R.L. Repeated batches as a strategy for high 2G ethanol production from undetoxified hemicellulose hydrolysate using immobilized cells of recombinant Saccharomyces cerevisiae in a fixed-bed reactor. Biotechnol. Biofuels 2020, 13, 85. [Google Scholar] [CrossRef]
- Alriksson, B.; Horváth, I.S.; Jönsson, L.J. Overexpression of Saccharomyces cerevisiae transcription factor and multidrug resistance genes conveys enhanced resistance to lignocellulose-derived fermentation inhibitors. Process Biochem. 2010, 45, 264–271. [Google Scholar] [CrossRef]
- Sürmeli, Y.; Holyavkin, C.; Topaloğlu, A.; Arslan, M.; Kısakesen, H.İ.; Çakar, Z.P. Evolutionary engineering and molecular characterization of a caffeine-resistant Saccharomyces cerevisiae strain. World J. Microbiol. 2019, 35, 183. [Google Scholar] [CrossRef] [PubMed]
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16. [Google Scholar] [CrossRef]
- Hong, K.K.; Vongsangnak, W.; Vemuri, G.N.; Nielsen, J. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. Proc. Natl. Acad. Sci. USA 2011, 19, 12179–12184. [Google Scholar] [CrossRef] [PubMed]
- Avrahami-Moyal, L.; Engelberg, D.; Wenger, J.W.; Sherlock, G.; Braun, S. Turbidostat culture of Saccharomyces cerevisiae W303-1A under selective pressure elicited by ethanol selects for mutations in SSD1 and UTH1. FEMS Yeast Res. 2012, 12, 521–533. [Google Scholar] [CrossRef]
- De Valk, S.C.; Bouwmeester, S.E.; de Hulster, E.; Mans, R. Engineering proton-coupled hexose uptake in Saccharomyces cerevisiae for improved ethanol yield. Biotechnol. Biofuels Bioprod. 2022, 15, 47. [Google Scholar] [CrossRef]
- Guadalupe-Medina, V.; Metz, B.; Oud, B.; van Der Graaf, C.M.; Mans, R.; Pronk, J.T.; van Maris, A.J. Evolutionary engineering of a glycerol-3-phosphate dehydrogenase-negative, acetate-reducing Saccharomyces cerevisiae strain enables anaerobic growth at high glucose concentrations. Microb. Biotechnol. 2014, 7, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kong, M.; Li, X.; Li, Q.; Xue, Q.; Hou, J.; Jia, Z.; Lei, Z.; Xiao, W.; Shi, S.; et al. Metabolic and evolutionary engineering of diploid yeast for the production of first-and second-generation ethanol. Front. Bioeng. Biotechnol. 2022, 9, 835928. [Google Scholar] [CrossRef]
- Dos Santos, V.L.; Carazzolle, M.F.; Nagamatsu, S.T.; Sampaio, N.M.V.; Almeida, L.D.; Pirolla, R.A.S.; Borelli, G.; Corrêa, T.L.R.; Argueso, J.L.; Pereira, G.A.G. Unraveling the genetic basis of xylose consumption in engineered Saccharomyces cerevisiae strains. Sci. Rep. 2016, 6, 38676. [Google Scholar] [CrossRef]
- Demeke, M.M.; Dietz, H.; Li, Y.; Foulquié-Moreno, M.R.; Mutturi, S.; Deprez, S.; Den Abt, T.; Bonini, B.M.; Liden, G.; Dumortier, F.; et al. Development of a D-xylose fermenting and inhibitor tolerant industrial Saccharomyces cerevisiae strain with high performance in lignocellulose hydrolysates using metabolic and evolutionary engineering. Biotechnol. Biofuels 2013, 6, 89. [Google Scholar] [CrossRef] [PubMed]
- Apel, A.R.; Ouellet, M.; Szmidt-Middleton, H.; Keasling, J.D.; Mukhopadhyay, A. Evolved hexose transporter enhances xylose uptake and glucose/xylose co-utilization in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 19512. [Google Scholar] [CrossRef] [PubMed]
- Turanlı-Yıldız, B.; Benbadis, L.; Alkım, C.; Sezgin, T.; Akşit, A.; Gökçe, A.; Öztürk, Y.; Baykal, A.T.; Cakar, Z.P.; Francois, J.M. In vivo evolutionary engineering for ethanol-tolerance of Saccharomyces cerevisiae haploid cells triggers diploidization. J. Biosci. Bioeng. 2017, 124, 309–318. [Google Scholar] [CrossRef]
- Mavrommati, M.; Papanikolaou, S.; Aggelis, G. Improving ethanol tolerance of Saccharomyces cerevisiae through adaptive laboratory evolution using high ethanol concentrations as a selective pressure. Process Biochem. 2023, 124, 280–289. [Google Scholar] [CrossRef]
- Liu, Z.L.; Slininger, P.J.; Gorsich, S.W. Enhanced biotransformation of furfural and hydroxymethylfurfural by newly developed ethanologenic yeast strains. Appl. Biochem. Biotechnol. 2005, 121, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.L.; Ma, M. Pathway-based signature transcriptional profiles as tolerance phenotypes for the adapted industrial yeast Saccharomyces cerevisiae resistant to furfural and HMF. Appl. Microbiol. Biotechnol. 2020, 104, 3473–3492. [Google Scholar] [CrossRef]
- Hacısalihoğlu, B.; Holyavkin, C.; Topaloğlu, A.; Kısakesen, H.İ.; Cakar, Z.P. Genomic and transcriptomic analysis of a coniferyl aldehyde-resistant Saccharomyces cerevisiae strain obtained by evolutionary engineering. FEMS Yeast Res. 2019, 19, foz021. [Google Scholar] [CrossRef]
- Salas-Navarrete, P.C.; de Oca Miranda, A.I.; Martínez, A.; Caspeta, L. Evolutionary and reverse engineering to increase Saccharomyces cerevisiae tolerance to acetic acid, acidic pH, and high temperature. Appl. Microbiol. Biotechnol. 2022, 106, 383–399. [Google Scholar] [CrossRef]
- Wallace-Salinas, V.; Gorwa-Grauslund, M.F. Adaptive evolution of an industrial strain of Saccharomyces cerevisiae for combined tolerance to inhibitors and temperature. Biotechnol. Biofuels 2013, 6, 151. [Google Scholar] [CrossRef]
- Wallace-Salinas, V.; Brink, D.P.; Ahrén, D.; Gorwa-Grauslund, M.F. Cell periphery-related proteins as major genomic tar-gets behind the adaptive evolution of an industrial Saccharomyces cerevisiae strain to combined heat and hydrolysate stress. BMC Genom. 2015, 16, 514. [Google Scholar] [CrossRef] [PubMed]
- Çakar, Z.P.; Seker, U.O.; Tamerler, C.; Sonderegger, M.; Sauer, U. Evolutionary engineering of multiple-stress resistant Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 569–578. [Google Scholar] [CrossRef]
- Kocaefe-Özşen, N.; Yilmaz, B.; Alkım, C.; Arslan, M.; Topaloğlu, A.; Gülsev, E.; Çakar, Z.P. Physiological and molecular characterization of an oxidative stress-resistant Saccharomyces cerevisiae strain obtained by evolutionary engineering. Front. Microbiol. 2022, 13, 822864. [Google Scholar] [CrossRef]
- Terzioğlu, E.; Alkım, C.; Arslan, M.; Balaban, B.G.; Holyavkin, C.; Kısakesen, H.İ.; Topaloğlu, A.; Yılmaz Şahin, Ü.; Gündüz Işık, S.; Akman, S.; et al. Genomic, transcriptomic and physiological analyses of silver-resistant Saccharomyces cerevisiae obtained by evolutionary engineering. Yeast 2020, 37, 413–426. [Google Scholar] [CrossRef] [PubMed]
- Arslan, M.; Holyavkin, C.; Kısakesen, H.İ.; Topaloğlu, A.; Sürmeli, Y.; Çakar, Z.P. Physiological and transcriptomic analysis of a chronologically long-lived Saccharomyces cerevisiae strain obtained by evolutionary engineering. Mol. Biotechnol. 2018, 60, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Holyavkin, C.; Turanlı-Yıldız, B.; Yılmaz, Ü.; Alkım, C.; Arslan, M.; Topaloğlu, A.; Kısakesen, H.İ.; de Billerbeck, G.; François, J.M.; Çakar, Z.P. Genomic, transcriptomic, and metabolic characterization of 2-Phenylethanol-resistant Saccharomyces cerevisiae obtained by evolutionary engineering. Front. Microbiol. 2023, 14, 1148065. [Google Scholar] [CrossRef]
- Yang, K.M.; Lee, N.R.; Woo, J.M.; Choi, W.; Zimmermann, M.; Blank, L.M.; Park, J.B. Ethanol reduces mitochondrial membrane integrity and thereby impacts carbon metabolism of Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 675–684. [Google Scholar] [CrossRef]
- Piotrowski, J.S.; Zhang, Y.; Bates, D.M.; Keating, D.H.; Sato, T.K.; Ong, I.M.; Landick, R. Death by a thousand cuts: The challenges and diverse landscape of lignocellulosic hydrolysate inhibitors. Front. Microbiol. 2014, 5, 90. [Google Scholar] [CrossRef] [PubMed]
- Ansanay-Galeote, V.; Blondin, B.; Dequin, S.; Sablayrolles, J.M. Stress effect of ethanol on fermentation kinetics by station-ary-phase cells of Saccharomyces cerevisiae. Biotechnol. Lett. 2001, 23, 677–681. [Google Scholar] [CrossRef]
- Adeboye, P.T.; Bettiga, M.; Olsson, L. The chemical nature of phenolic compounds determines their toxicity and induces distinct physiological responses in Saccharomyces cerevisiae in lignocellulose hydrolysates. AMB Express 2014, 4, 46. [Google Scholar] [CrossRef] [PubMed]
- Wenger, J.W.; Piotrowski, J.; Nagarajan, S.; Chiotti, K.; Sherlock, G.; Rosenzweig, F. Hunger artists: Yeast adapted to carbon limitation show trade-offs under carbon sufficiency. PLoS Genet. 2011, 7, e1002202. [Google Scholar] [CrossRef] [PubMed]
- Caspeta, L.; Nielsen, J. Thermotolerant yeast strains adapted by laboratory evolution show trade-off at ancestral temperatures and preadaptation to other stresses. MBio 2015, 6, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Mans, R.; Daran, J.M.; Pronk, J.T. Under pressure: Evolutionary engineering of yeast strains for improved performance in fuels and chemicals production. Curr. Opin. Biotechnol. 2018, 50, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Wisselink, H.W.; Toirkens, M.J.; Wu, Q.; Pronk, J.T.; van Maris, A.J. Novel evolutionary engineering approach for accelerated utilization of glucose, xylose, and arabinose mixtures by engineered Saccharomyces cerevisiae strains. Appl. Environ. Microbiol. 2009, 75, 907–914. [Google Scholar] [CrossRef]
- Radek, A.; Tenhaef, N.; Müller, M.F.; Brüsseler, C.; Wiechert, W.; Marienhagen, J.; Polen, T.; Noack, S. Miniaturized and automated adaptive laboratory evolution: Evolving Corynebacterium glutamicum towards an improved D-xylose utilization. Bioresour. Technol. 2017, 245, 1377–1385. [Google Scholar] [CrossRef]
- Wang, J.; Jian, X.; Xing, X.H.; Zhang, C.; Fei, Q. Empowering a methanol-dependent Escherichia coli via adaptive evolution using a high-throughput microbial microdroplet culture system. Front. Bioeng. Biotechnol. 2020, 8, 570. [Google Scholar] [CrossRef] [PubMed]
- Ekkers, D.M.; Branco dos Santos, F.; Mallon, C.A.; Bruggeman, F.; van Doorn, G.S. The omnistat: A flexible continuous-culture system for prolonged experimental evolution. Methods Ecol. Evol. 2020, 11, 932–942. [Google Scholar] [CrossRef] [PubMed]
- LaCroix, R.A.; Palsson, B.O.; Feist, A.M. A model for designing adaptive laboratory evolution experiments. Appl. Environ. Microbiol. 2017, 83, 03115–03116. [Google Scholar] [CrossRef] [PubMed]
- Wong, B.G.; Mancuso, C.P.; Kiriakov, S.; Bashor, C.J.; Khalil, A.S. Precise, automated control of conditions for high-throughput growth of yeast and bacteria with eVOLVER. Nat. Biotechnol. 2018, 36, 614–623. [Google Scholar] [CrossRef]
- LaCroix, R.A.; Sandberg, T.E.; O’Brien, E.J.; Utrilla, J.; Ebrahim, A.; Guzman, G.I.; Szubin, R.; Palsson, B.O.; Feist, A.M. Use of adaptive laboratory evolution to discover key mutations enabling rapid growth of Escherichia coli K-12 MG1655 on glucose minimal medium. Appl. Environ. Microbiol. 2015, 81, 17–30. [Google Scholar] [CrossRef]
- Winkler, J.D.; Kao, K.C. Recent advances in the evolutionary engineering of industrial biocatalysts. Genomics 2014, 104, 406–411. [Google Scholar] [CrossRef]
- Amer, B.; Baidoo, E.E. Omics-driven biotechnology for industrial applications. Front. Bioeng. Biotechnol. 2021, 9, 613307. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Xia, S.; Dong, C.; Pan, H.; Cai, J.; Huang, L.; Xu, Z.; Lian, J. Random base editing for genome evolution in Saccharomyces cerevisiae. ACS Synth. Biol. 2021, 20, 2440–2446. [Google Scholar] [CrossRef] [PubMed]
- Cao, M.; Tran, V.G.; Zhao, H. Unlocking nature’s biosynthetic potential by directed genome evolution. Curr. Opin. Biotechnol. 2020, 66, 95–104. [Google Scholar] [CrossRef]
- Alper, H.; Stephanopoulos, G. Global transcription machinery engineering: A new approach for improving cellular phenotype. Metab. Eng. 2007, 9, 258–267. [Google Scholar] [CrossRef]
- Ke, T.; Liu, J.; Zhao, S.; Wang, X.; Wang, L.; Li, Y.; Lu, Y.; Hui, F. Using global transcription machinery engineering (GTME) and site-saturation mutagenesis technique to improve ethanol yield of Saccharomyces cerevisiae. Appl. Biochem. Microbiol. 2020, 56, 563–568. [Google Scholar] [CrossRef]
- Xue, T.; Chen, D.; Su, Q.; Yuan, X.; Liu, K.; Huang, L.; Fang, J.; Chen, J.; He, W.; Chen, Y. Improved ethanol tolerance and production of Saccharomyces cerevisiae by global transcription machinery engineering via directed evolution of the SPT8 gene. Food Biotechnol. 2019, 3, 155–173. [Google Scholar] [CrossRef]
- Xue, T.; Liu, K.; Chen, D.; Yuan, X.; Fang, J.; Yan, H.; Huang, L.; Chen, Y.; He, W. Improved bioethanol production using CRISPR/Cas9 to disrupt the ADH2 gene in Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2018, 34, 154. [Google Scholar] [CrossRef]
- Claes, A.; Deparis, Q.; Foulquié-Moreno, M.R.; Thevelein, J.M. Simultaneous secretion of seven lignocellulolytic enzymes by an industrial second-generation yeast strain enables efficient ethanol production from multiple polymeric substrates. Metab. Eng. 2020, 59, 131–141. [Google Scholar] [CrossRef]
- Perez, C.L.; Milessi, T.S.; Sandri, J.P.; Ramos, M.D.; Carvalho, B.T.; Claes, A.; Demeke, M.M.; Thevelein, J.M.; Zangirolami, T.C. Evaluation of Consolidated Bioprocessing of Sugarcane Biomass by a Multiple Hydrolytic Enzyme Producer Saccharomyces Yeast. Bioenerg. Res. 2023. [Google Scholar] [CrossRef]
- Yang, P.; Jiang, S.; Lu, S.; Jiang, S.; Jiang, S.; Deng, Y.; Lu, J.; Wang, H.; Zhou, Y. Ethanol yield improvement in Saccharomyces cerevisiae GPD2 Delta FPS1 Delta ADH2 Delta DLD3 Delta mutant and molecular mechanism exploration based on the metabolic flux and transcriptomics approaches. Microb. Cell Factories 2022, 21, 160. [Google Scholar] [CrossRef] [PubMed]
- Jakočiūnas, T.; Jensen, M.K.; Keasling, J.D. CRISPR/Cas9 advances engineering of microbial cell factories. Metab. Eng. 2016, 34, 44–59. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.H.; Isaacs, F.J.; Carr, P.A.; Sun, Z.Z.; Xu, G.; Forest, C.R.; Church, G.M. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009, 13, 894–898. [Google Scholar] [CrossRef] [PubMed]
- Si, T.; Chao, R.; Min, Y.; Wu, Y.; Ren, W.; Zhao, H. Automated multiplex genome-scale engineering in yeast. Nat. Commun. 2017, 4, 15187. [Google Scholar] [CrossRef]
- Momen-Roknabadi, A.; Oikonomou, P.; Zegans, M.; Tavazoie, S. An inducible CRISPR interference library for genetic interrogation of Saccharomyces cerevisiae biology. Commun. Biol. 2020, 27, 723. [Google Scholar] [CrossRef] [PubMed]



| Purpose | Modification | Improvement | Reference | |
|---|---|---|---|---|
| Lowering ATP yield | Introduction of futile cycle | Expression of E. coli FBPase insensitive to fructose-2,6-bisphosphate inhibition | 8.8% higher bioethanol yield | [44] |
| Introduction of ATPase enzyme | Heterologous expression of the F1 part of E. coli H+-ATPase enzyme | 10% higher bioethanol yield | [45] | |
| Increasing the unspecific alkaline phosphatase activity | Overexpression of PHO8 gene | 13% higher bioethanol yield | [46] | |
| Introduction of Entner-Doudoroff (ED) pathway | Expression of KDPG (2-keto-3-deoxy-6-phosphogluconate) aldolase from E. coli | - | [48] | |
| Relocation of sucrose hydrolysis from the extracellular space to the cytosol | Engineering the promoter and 5′ coding sequences of SUC2 gene | 4% higher bioethanol yield | [17] | |
| Sustainable reduction of glycerol formation | Fine-tuning of glycerol 3-phosphate dehydrogenases (GPDH) | Introducing lower-strength TEF1 promoters to GPD1 and GPD2 genes | 5% higher bioethanol yield | [54] |
| Fine-tuning of glycerol 3-phosphate dehydrogenase | Engineering the promoter of GPD1 in a gpd2Δ background | 7% higher bioethanol yield | [55] | |
| Introduction of direct cofactor regulation strategies | Expressing B. cereus gapN gene, E. coli frdA gene and mhpF gene independently | Increased bioethanol yield | [57] | |
| Engineering of ammonium assimilation | Overexpression of GLN1 and GLT1 genes, deletion of GDH1 gene | 10% higher bioethanol yield | [58] | |
| Introduction of Calvin-cycle enzymes | Deletion of GPD2 and heterologous expression of PRK and RuBisCO | 15% higher bioethanol yield | [59] | |
| Prevention of bacterial contamination | Introduction of phage endolysin on the cell surface | Anchoring recombinant peptidoglycan hydrolase, endolysin LysKB317, by using cell surface display | 83.8% decrease in bacterial cell counts | [72] |
| Introduction of xylose catabolism | Construction of xylose reductase-xylitol dehydrogenase (XR-XDH) pathway | Genomic integration of XYL1 and XYL2 genes from S. stipitis, XKS1 from S. cerevisiae, BGL1 from A. aculeatus, and GXS1 from C. intermedia | Xylose consumption rate of 6.62 g/L/h and an ethanol yield of 0.394 | [80] |
| Construction of xylose reductase-xylitol dehydrogenase (XR-XDH) pathway | Expression of XYL1.2 from S. passalidarum and XYL2 from S. stipitis | 0.40 g g−1 CDW ethanol yield and 0.33 g g−1 CDW h−1 productivity | [81] | |
| Construction of xylose isomerase (XI) pathway | Expression of XI gene (xylA) from the bacterium B. cenocepacia | 5-fold increase in xylose consumption and over 1.5-fold increase in ethanol production | [83] | |
| Construction of xylose isomerase (XI) pathway | Expression of XI from the bacterium P. acidipropionici with the co-expression of GroEL-GroES chaperonin complex from E. coli | Yield of 0.44 g ethanol/g xylose | [85] | |
| Improving xylose catabolism | Lowering xylitol production | Deletion of GRE3 gene in S. cerevisiae | Increased ethanol yield of 0.47 g/g of total sugars during fermentation of corn-cob hydrolysate | [79] |
| Introduction of a synthetic reductive PPP for carbon dioxide recycling | Expression of RuBisCO and PRK enzymes in a S. cerevisiae strain harboring the XR/XDH pathway | Increased ethanol yield and reduced release of carbon dioxide | [82] | |
| Increasing accessibility of lignocellulosic biomass | Conversion of cellulose into glucose | Expression of cassette carrying a cellulase gene from A. gigas Spix | 37.7-fold higher ethanol yield | [88] |
| Increasing stress tolerance | Increasing osmotolerance | Overexpression of TPS1 and TPS2 genes in S. cerevisiae | Increased ethanol yield and osmotolerance, decreased glycerol production | [61] |
| Increasing thermotolerance | Overexpression of RSP5 gene | Thermotolerance at 41 °C and ability to tolerate higher temperatures. | [64] | |
| Increasing ethanol tolerance | Overexpression of a truncated version of the MSN2 gene in an industrial fuel ethanol strain | Increased ethanol tolerance (up to 14%) | [66] | |
| Increasing acetate tolerance | Overexpression of HAA1 gene | The addition of acetate at 0.5% (w/v, pH 4.5) does not inhibit ethanol production | [71] | |
| Increasing HMF tolerance | Overexpression of ADH1 and ADH6 genes | Higher specific ethanol productivity in the presence of HMF | [91] | |
| Increasing acetic acid tolerance | Deletion of ADY2 gene | 14.7% increase in ethanol yield, in the presence of 3.6 g/L acetic acid | [94] | |
| Increasing coniferyl aldehyde tolerance | Overexpression of ATR1 and FLR1 genes | Increased coniferyl aldehyde tolerance | [96] | |
| Purpose | Modification | Improvement and/or the Associated Mutations/Changes Detected | Reference | |
|---|---|---|---|---|
| Increasing Growth Rate and Viability | Faster growth and galactose utilization | RAS2 mutation detected | 24% increased specific growth rate on galactose and higher ethanol yield | [99] |
| Improved growth rate under ethanol stress | SSD1 and UTH1 mutations detected | Increased specific growth rate from 0.029 h−1 to 0.32 h−1 at 8% (v/v) ethanol | [100] | |
| Decreasing By-Product Formation | Decreased biomass formation | Replacement of diffusion mediated hexose transporters with a proton-coupled transport system | 44–47.6% decreased biomass production and 17.2% increased ethanol yield | [101] |
| Decreased glycerol production | Evolutionary engineering of a gpd1Δ and gpd2Δ S. cerevisiae strain expressing mhpF from E. coli for osmotolerance revealed the mutation mhpF D38N | Increased ethanol yield from 79% (reference) to 92%, and lower glycerol production (0.64 g/L) | [102] | |
| Decreased glycerol production | Evolutionary engineering in 15% wheat straw hydrolysate of an industrial yeast strain incorporating xylose genes | 63.1% ethanol yield from cellulose and xylose, and 20% lower glycerol production | [103] | |
| Improving Utilization and Transport of Sugars | Improved xylose utilization | ISU1 and SSK2 mutations detected | Improved yield of 0.46 g ethanol/xylose | [104] |
| Improved xylose utilization | Evolutionary engineering of a S. cerevisiae strain expressing C. phytofermentans XylA gene encoding XI and genes encoding PPP enzymes | Improved maximum specific xylose consumption rate of 1.1 g/g CDW/h in synthetic medium, and 32% higher ethanol production | [105] | |
| Improved xylose utilization | HXT7 mutation detected | Improved xylose uptake rate (Vmax = 186.4 nmol min−1mg−1) | [106] | |
| Increasing Tolerance to Ethanol and Lignocellulosic Inhibitors | Increased ethanol resistance | Triggering of diploidization | Resistance against 12% (v/v) ethanol stress | [107] |
| Increased ethanol resistance | Increased glucose uptake rate and decreased lag phase | Resistance against 25% ethanol for 4 h | [108] | |
| Increased Furfural and HMF resistance | Transcriptomic changes associated with Yap1, Met4, Msn2/4 and Pdr1/3 transcription factors detected | 30 mM furfural and 60 mM HMF resistance without loss of ethanol yield | [109,110] | |
| Increased Coniferyl aldehyde resistance | PDR1, GLN3 and CRZ1 mutations detected | Resistance against 2 mM coniferyl aldehyde | [111] | |
| Increased Thermo-acid tolerance | RAS2 and HSF1 mutations detected | Tolerance to 12 g/L acetate at pH 4 and 30 °C | [112] | |
| Increased thermotolerance | MTL1, FLO9/FLO11, and CYC3 gene mutations detected | Furfural and HMF tolerance under thermal stress (39 °C) | [113,114] | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Topaloğlu, A.; Esen, Ö.; Turanlı-Yıldız, B.; Arslan, M.; Çakar, Z.P. From Saccharomyces cerevisiae to Ethanol: Unlocking the Power of Evolutionary Engineering in Metabolic Engineering Applications. J. Fungi 2023, 9, 984. https://doi.org/10.3390/jof9100984
Topaloğlu A, Esen Ö, Turanlı-Yıldız B, Arslan M, Çakar ZP. From Saccharomyces cerevisiae to Ethanol: Unlocking the Power of Evolutionary Engineering in Metabolic Engineering Applications. Journal of Fungi. 2023; 9(10):984. https://doi.org/10.3390/jof9100984
Chicago/Turabian StyleTopaloğlu, Alican, Ömer Esen, Burcu Turanlı-Yıldız, Mevlüt Arslan, and Zeynep Petek Çakar. 2023. "From Saccharomyces cerevisiae to Ethanol: Unlocking the Power of Evolutionary Engineering in Metabolic Engineering Applications" Journal of Fungi 9, no. 10: 984. https://doi.org/10.3390/jof9100984
APA StyleTopaloğlu, A., Esen, Ö., Turanlı-Yıldız, B., Arslan, M., & Çakar, Z. P. (2023). From Saccharomyces cerevisiae to Ethanol: Unlocking the Power of Evolutionary Engineering in Metabolic Engineering Applications. Journal of Fungi, 9(10), 984. https://doi.org/10.3390/jof9100984