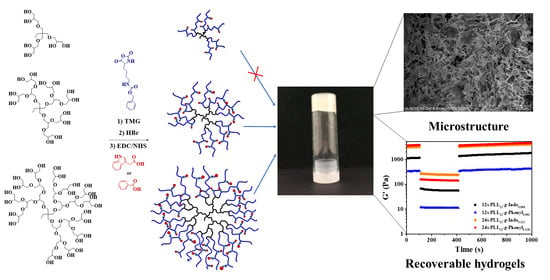
Synthesis and Hydrogelation of Star-Shaped Graft Copolypetides with Asymmetric Topology
Abstract
:1. Introduction
2. Results and Discussion
2.1. Star-Shaped Graft Copolypeptide Synthesis and Characterization
2.2. Critical Gelation Concentrations (CGCs) and Hydrogelation of Star-Shaped Graft Copolypeptides
2.3. Secondary Structure of Star-Shaped Graft Copolypeptides
2.4. Self-Assembled Nanostructure and Gel Morphology of Star-Shaped Graft Copolypeptides
2.5. Mechanical Strength and Recovery Behavior of Star-Shaped Graft Copolypeptides
3. Conclusions
4. Materials and Methods
4.1. Materials
4.2. Synthesis and Characterization of Star-Shaped Poly(Z-L-lysine) Polypeptides (s-PZLL)
4.3. Deprotection of Z-Group on Polypeptides
4.4. Synthesis and Characterization of Star-Shaped Poly(L-lysine) Graft Copolypeptides
4.5. Preparation of Star-Shaped Graft Copolypeptide Hydrogel Samples and Determination of Critical Gelation Concentrations (CGCs)
4.6. Determination of Secondary Structure Adoption
4.7. Characterization of Graft Copolypeptide Hydrogel Self-Assembled Nanostructures
4.8. Characterization of Mechanical Strength and Recovery Behavior
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fu, J. Hydrogel properties and applications. J. Mater. Chem. B 2019, 7, 1523–1525. [Google Scholar] [CrossRef] [PubMed]
- Oyen, M. Mechanical characterisation of hydrogel materials. Int. Mater. Rev. 2014, 59, 44–59. [Google Scholar] [CrossRef]
- Vedadghavami, A.; Minooei, F.; Mohammadi, M.H.; Khetani, S.; Kolahchi, A.R.; Mashayekhan, S.; Sanati-Nezhad, A. Manufacturing of hydrogel biomaterials with controlled mechanical properties for tissue engineering applications. Acta Biomater. 2017, 62, 42–63. [Google Scholar] [CrossRef] [PubMed]
- Chyzy, A.; Plonska-Brzezinska, M.E. Hydrogel properties and their impact on regenerative medicine and tissue engineering. Molecules 2020, 25, 5795. [Google Scholar] [CrossRef]
- Saini, K. Preparation method, Properties and Crosslinking of hydrogel: A review. PharmaTutor 2017, 5, 27–36. [Google Scholar]
- Darge, H.F.; Andrgie, A.T.; Tsai, H.-C.; Lai, J.-Y. Polysaccharide and polypeptide based injectable thermo-sensitive hydrogels for local biomedical applications. Int. J. Biol. Macromol. 2019, 133, 545–563. [Google Scholar] [CrossRef]
- Hamidi, M.; Azadi, A.; Rafiei, P. Hydrogel nanoparticles in drug delivery. Adv. Drug Del. Rev. 2008, 60, 1638–1649. [Google Scholar] [CrossRef]
- Deligkaris, K.; Tadele, T.S.; Olthuis, W.; van den Berg, A. Hydrogel-based devices for biomedical applications. Sens. Actuators B Chem. 2010, 147, 765–774. [Google Scholar] [CrossRef]
- Thakur, S.; Thakur, V.K.; Arotiba, O.A. History, classification, properties and application of hydrogels: An overview. Hydrogels 2018, 29–50. [Google Scholar] [CrossRef]
- Van Bemmelen, J. Das hydrogel und das krystallinische hydrat des kupferoxyds. Z. Anorg. Chem. 1894, 5, 466–483. [Google Scholar] [CrossRef] [Green Version]
- Wichterle, O.; Lim, D. Hydrophilic gels for biological use. Nature 1960, 185, 117–118. [Google Scholar] [CrossRef]
- Zhang, L.; Furst, E.M.; Kiick, K.L. Manipulation of hydrogel assembly and growth factor delivery via the use of peptide–polysaccharide interactions. J. Control. Release 2006, 114, 130–142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, R.; Li, S.; Zhang, N.; Shen, G.; Mohwald, H.; Yan, X. Self-assembled injectable peptide hydrogels capable of triggering antitumor immune response. Biomacromolecules 2017, 18, 3514–3523. [Google Scholar] [CrossRef]
- Li, S.; Xing, R.; Chang, R.; Zou, Q.; Yan, X. Nanodrugs based on peptide-modulated self-assembly: Design, delivery and tumor therapy. Curr. Opin. Colloid Interface Sci. 2018, 35, 17–25. [Google Scholar] [CrossRef]
- Kiick, K.L. Peptide-and protein-mediated assembly of heparinized hydrogels. Soft Matter 2008, 4, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Bonduelle, C. Secondary structures of synthetic polypeptide polymers. Polym. Chem. 2018, 9, 1517–1529. [Google Scholar] [CrossRef]
- Chen, C.; Lan, J.; Li, Y.; Liang, D.; Ni, X.; Liu, Q. Secondary structure-governed polypeptide cross-linked polymeric hydrogels. Chem. Mater. 2019, 32, 1153–1161. [Google Scholar] [CrossRef]
- Fan, J.; Li, R.; Wang, H.; He, X.; Nguyen, T.P.; Letteri, R.A.; Zou, J.; Wooley, K.L. Multi-responsive polypeptide hydrogels derived from N-carboxyanhydride terpolymerizations for delivery of nonsteroidal anti-inflammatory drugs. Org. Biomol. Chem. 2017, 15, 5145–5154. [Google Scholar] [CrossRef]
- Zhao, S.; Zhu, H.; Chen, Z.; Shuai, S.; Zhang, N.; Liu, Y.; Rao, Z.; Li, Y.; Zhao, C.; Zhou, K. Preparation and properties of a temperature-and pH-responsive polypeptide hydrogel. Mater. Res. Express 2019, 6, 085711. [Google Scholar] [CrossRef]
- Lin, J.-Y.; Lai, P.-L.; Lin, Y.-K.; Peng, S.; Lee, L.-Y.; Chen, C.-N.; Chu, I.-M. A poloxamer-polypeptide thermosensitive hydrogel as a cell scaffold and sustained release depot. Polym. Chem. 2016, 7, 2976–2985. [Google Scholar] [CrossRef]
- Huang, C.-C.; Phan, T.H.M.; Ooya, T.; Kawasaki, S.; Lin, B.-Y.; Jan, J.-S. Effect of tethered sheet-like motif and asymmetric topology on hydrogelation of star-shaped block copolypeptides. Polymer 2022, 250, 124864. [Google Scholar] [CrossRef]
- Phan, T.H.M.; Huang, C.-C.; Tsai, Y.-J.; Hu, J.-J.; Jan, J.-S. Polypeptide Composition and Topology Affect Hydrogelation of Star-Shaped Poly (L-lysine)-Based Amphiphilic Copolypeptides. Gels 2021, 7, 131. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-Y.; Tang, C.-C.; Jan, J.-S. Synthesis and hydrogelation of star-shaped poly (L-lysine) polypeptides modified with different functional groups. Polymer 2018, 151, 108–116. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, S.; Wan, Y.; Fu, W.; Li, Z. Hydrogels assembled from star-shaped polypeptides with a dendrimer as the core. Soft Matter 2015, 11, 2945–2951. [Google Scholar] [CrossRef]
- Murphy, R.; Borase, T.; Payne, C.; O’Dwyer, J.; Cryan, S.-A.; Heise, A. Hydrogels from amphiphilic star block copolypeptides. RSC Adv. 2016, 6, 23370–23376. [Google Scholar] [CrossRef]
- Lu, D.; Li, Y.; Wang, X.; Zhang, Y.; Guo, H.; Sun, S.; Wang, X.; Zhang, Y.; Lei, Z. All-in-one hyperbranched polypeptides for surgical adhesives and interventional embolization of tumors. J. Mater. Chem. B 2018, 6, 7511–7520. [Google Scholar] [CrossRef]
- Tang, C.-C.; Zhang, S.-H.; Phan, T.H.M.; Tseng, Y.-C.; Jan, J.-S. Block length and topology affect self-assembly and gelation of poly (L-lysine)-block-poly (S-benzyl-l-cysteine) block copolypeptides. Polymer 2021, 228, 123891. [Google Scholar] [CrossRef]
- Chen, Y.-F.; Lai, Y.-D.; Chang, C.-H.; Tsai, Y.-C.; Tang, C.-C.; Jan, J.-S. Star-shaped polypeptides exhibit potent antibacterial activities. Nanoscale 2019, 11, 11696–11708. [Google Scholar] [CrossRef]
- Wang, Y.; Chang, Y.C. Synthesis and conformational transition of surface-tethered polypeptide: Poly (L-lysine). Macromolecules 2003, 36, 6511–6518. [Google Scholar] [CrossRef]
- St. Pierre, S.; Ingwall, R.; Verlander, M.; Goodman, M. Conformational studies of sequential polypeptides containing lysine and tyrosine. Biopolym. Orig. Res. Biomol. 1978, 17, 1837–1848. [Google Scholar] [CrossRef]
- Stanojlovic, V.; Müller, A.; Moazzam, A.; Hinterholzer, A.; Ożga, K.; Berlicki, Ł.; Schubert, M.; Cabrele, C. A Conformationally Stable Acyclic β-Hairpin Scaffold Tolerating the Incorporation of Poorly β-Sheet-Prone Amino Acids. ChemBioChem 2022, 23, e202100604. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Mahalakshmi, R.; Shamala, N.; Balaram, P. Aromatic interactions in tryptophan-containing peptides: Crystal structures of model tryptophan peptides and phenylalanine analogs. J. Pept. Res. 2005, 65, 113–129. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, R.; Sengupta, A.; Raghothama, S.; Shamala, N.; Balaram, P. Tryptophan rich peptides: Influence of indole rings on backbone conformation. Pept. Sci. Orig. Res. Biomol. 2007, 88, 36–54. [Google Scholar] [CrossRef]
- Sun, Y.; Wollenberg, A.L.; O’Shea, T.M.; Cui, Y.; Zhou, Z.H.; Sofroniew, M.V.; Deming, T.J. Conformation-directed formation of self-healing diblock copolypeptide hydrogels via polyion complexation. J. Am. Chem. Soc. 2017, 139, 15114–15121. [Google Scholar] [CrossRef]
- Huang, J.; Hastings, C.L.; Duffy, G.P.; Kelly, H.M.; Raeburn, J.; Adams, D.J.; Heise, A. Supramolecular hydrogels with reverse thermal gelation properties from (oligo) tyrosine containing block copolymers. Biomacromolecules 2013, 14, 200–206. [Google Scholar] [CrossRef]
- Liu, Y.; Zhan, G.; Zhong, X.; Yu, Y.; Gan, W. Effect of pi–pi stacking on the self-assembly of azomethine-type rod–coil liquid crystals. Liq. Cryst. 2011, 38, 995–1006. [Google Scholar] [CrossRef]
- Das, A. Studies on complex π-π and T-stacking features of imidazole and phenyl/p-halophenyl units in series of 5-amino-1-(phenyl/p-halophenyl) imidazole-4-carboxamides and their carbonitrile derivatives: Role of halogens in tuning of conformation. J. Mol. Struct. 2017, 1147, 520–540. [Google Scholar] [CrossRef]
- Chu, B. Laser Light Scattering: Basic Principles and Practice; Courier Corporation: Chelmsford, MA, USA, 2007. [Google Scholar]
- Tsai, Y.-L.; Tseng, Y.-C.; Chen, Y.-M.; Wen, T.-C.; Jan, J.-S. Zwitterionic polypeptides bearing carboxybetaine and sulfobetaine: Synthesis, self-assembly, and their interactions with proteins. Polym. Chem. 2018, 9, 1178–1189. [Google Scholar] [CrossRef]
- Beychok, S.; Fasman, G.D. Circular dichroism of poly-L-tyrosine. Biochemistry 1964, 3, 1675–1678. [Google Scholar] [CrossRef]
- Fasman, G.D.; Bodenheimer, E.; Lindblow, C. Optical rotatory dispersion studies of Poly-L-tyrosine and copolymers of L-glutamic acid and L-tyrosine. Significance of the tyrosyl Cotton effects with respect to protein conformation. Biochemistry 1964, 3, 1665–1674. [Google Scholar] [CrossRef]
- Hirschmann, R.; Schwam, H.; Strachan, R.; Schoenewaldt, E.; Barkemeyer, H.; Miller, S.; Conn, J.B.; Garsky, V.; Veber, D.F.; Denkewalter, R.G. Controlled synthesis of peptides in aqueous medium. VIII. Preparation and use of novel. alpha.-amino acid N-carboxyanhydrides. J. Am. Chem. Soc. 1971, 93, 2746–2754. [Google Scholar] [CrossRef] [PubMed]
- Ooya, T.; Ogawa, T.; Takeuchi, T. Temperature-induced recovery of a bioactive enzyme using polyglycerol dendrimers: Correlation between bound water and protein interaction. J. Biomater. Sci. Polym. Ed. 2018, 29, 701–715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.-S.; Hsu, Y.-Y.; Lin, J.-H.; Jan, J.-S. Alkyl-poly (L-threonine)/cyclodextrin supramolecular hydrogels with different molecular assemblies and gel properties. ACS Macro Lett. 2016, 5, 1201–1205. [Google Scholar] [CrossRef] [PubMed]
- Jommanee, N.; Chanthad, C.; Manokruang, K. Preparation of injectable hydrogels from temperature and pH responsive grafted chitosan with tuned gelation temperature suitable for tumor acidic environment. Carbohydr. Polym. 2018, 198, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Kitagawa, M.; Maeda, T.; Hotta, A. PEG-based nanocomposite hydrogel: Thermo-responsive sol-gel transition and degradation behavior controlled by the LA/GA ratio of PLGA-PEG-PLGA. Polym. Degrad. Stab. 2018, 147, 222–228. [Google Scholar] [CrossRef]






| Polypeptide | Grafting Ratio (%) | Grafting Efficiency (%) | CGC (wt%) | Secondary Structure | |
|---|---|---|---|---|---|
| Random Coil (%) | β-Sheet/β-Turn (%) | ||||
| 6s-PLL32-g-Indo0.08 | 7.9 | 26 | - * | 41.9 | 37.2/16.1 |
| 6s-PLL32-g-Phenyl0.08 | 7.9 | 26 | - * | 42.8 | 37.3/16.2 |
| 12s-PLL12-g-Indo0.10 | 9.8 | 33 | 5.85 | 43.1 | 36.7/16.8 |
| 12s-PLL12-g-Phenyl0.08 | 8.1 | 27 | 6.3 | 44.5 | 35.5/16.6 |
| 24s-PLL12-g-Indo0.11 | 11.2 | 37 | 2.9 | 42.6 | 39.1/15.4 |
| 24s-PLL12-g-Phenyl0.13 | 12.8 | 43 | 3.6 | 43.3 | 36.7/15.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Phan, T.H.M.; Yang, Y.-H.; Tsai, Y.-J.; Chung, F.-Y.; Ooya, T.; Kawasaki, S.; Jan, J.-S. Synthesis and Hydrogelation of Star-Shaped Graft Copolypetides with Asymmetric Topology. Gels 2022, 8, 366. https://doi.org/10.3390/gels8060366
Phan THM, Yang Y-H, Tsai Y-J, Chung F-Y, Ooya T, Kawasaki S, Jan J-S. Synthesis and Hydrogelation of Star-Shaped Graft Copolypetides with Asymmetric Topology. Gels. 2022; 8(6):366. https://doi.org/10.3390/gels8060366
Chicago/Turabian StylePhan, Thi Ha My, Yu-Hsun Yang, Yi-Jen Tsai, Fang-Yu Chung, Tooru Ooya, Shiho Kawasaki, and Jeng-Shiung Jan. 2022. "Synthesis and Hydrogelation of Star-Shaped Graft Copolypetides with Asymmetric Topology" Gels 8, no. 6: 366. https://doi.org/10.3390/gels8060366
APA StylePhan, T. H. M., Yang, Y. -H., Tsai, Y. -J., Chung, F. -Y., Ooya, T., Kawasaki, S., & Jan, J. -S. (2022). Synthesis and Hydrogelation of Star-Shaped Graft Copolypetides with Asymmetric Topology. Gels, 8(6), 366. https://doi.org/10.3390/gels8060366