MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2
Abstract
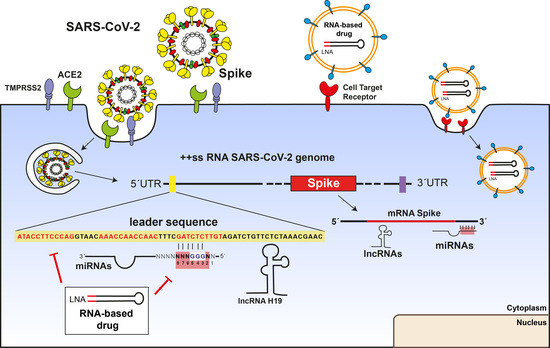
:1. Introduction
2. Materials and Methods
2.1. Dataset
2.2. miRNAs and lncRNAs Interaction with SARS-CoV-2 Genome
| IntaRNA |
| IntaRNA -t <input_file_query> -q <input_file_target> > <output_file> |
| RNAplex |
| RNAplfold -W <mirna_length> -u <mirna_length> -O --plex_output < <input_file> |
| RNAplex -l <mirna_length> -q <input_file_query> -t <input_file_target> -a ./ |
| RNAup |
| RNAup -w <mirna_length> -b -o -3 -5 --interaction_first < <input_file> |
2.3. RNA–RNA Interaction Analysis
2.4. MiRNAs and SARS-CoV-2 Leader Sequence Motifs Analysis
2.5. Logistic Prediction of miRNA-Target Sites Using High Throughput and V-CLIP Studies
2.6. LncRNA Secondary Structures
3. Results
3.1. MiRNAs Bind the 5′UTR-Leader Sequence, the 3′UTR of SARS-CoV-2 and the Spike mRNA through Noncanonical Bindings
3.2. MiRNAs and the Leader Sequence Contain Motifs Increasing miRNA: Viral RNA Selectivity
3.3. lncRNAs H19, LINC01505, and Fendrr Interact with SARS-CoV-2 and Spike mRNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Genet. 2019, 17, 181–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shereen, M.A.; Khan, S.; Kazmi, A.; Bashir, N.; Siddique, R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020, 24, 91–98. [Google Scholar] [CrossRef]
- Lai, C.-C.; Shih, T.-P.; Ko, W.-C.; Tang, H.-J.; Hsueh, P.-R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona-virus disease-2019 (COVID-19): The epidemic and the challenges. Int. J. Antimicrob. Agents. 2020, 55, 105924. [Google Scholar] [CrossRef]
- Vetter, P.; Kaiser, L.; Calmy, A.; Agoritsas, T.; Huttner, A. Dexamethasone and remdesivir: Finding method in the COVID-19 madness. Lancet Microbe 2020, 1, e309–e310. [Google Scholar] [CrossRef]
- Cohen, M.S. Hydroxychloroquine for the Prevention of Covid-19—Searching for Evidence. N. Engl. J. Med. 2020, 383, 585–586. [Google Scholar] [CrossRef]
- Geleris, J.; Sun, Y.; Platt, J.; Zucker, J.; Baldwin, M.; Hripcsak, G.; Labella, A.; Manson, D.K.; Kubin, C.; Barr, R.G.; et al. Observational Study of Hydroxychloroquine in Hospitalized Pa-tients with Covid-19. N. Engl. J. Med. 2020, 382, 2411–2418. [Google Scholar] [CrossRef] [PubMed]
- Poller, W.; Dimmeler, S.; Heymans, S.; Zeller, T.; Haas, J.; Karakas, M.; Leistner, D.-M.; Jakob, P.; Nakagawa, S.; Blankenberg, S.; et al. Non-coding RNAs in cardiovascular diseases: Diagnostic and therapeutic perspectives. Eur. Hear. J. 2018, 39, 2704–2716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Civenni, G. Targeting Promoter-Associated Noncoding RNA In Vivo. Methods Mol. Biol. 2017, 1543, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Bajan, S.; Hutvagner, G. RNA-Based Therapeutics: From Antisense Oligonucleotides to miRNAs. Cells 2020, 9, 137. [Google Scholar] [CrossRef] [Green Version]
- Lundstrom, K. RNA-based drugs and vaccines. Expert Rev. Vaccines 2015, 14, 253–263. [Google Scholar] [CrossRef]
- Natarelli, L.; Weber, C. Next-Generation Therapeutic Concepts for Atherosclerosis: Focus on Cell Specificity and Noncoding RNAs. Thromb. Haemost. 2019, 119, 1199–1201. [Google Scholar] [CrossRef]
- Ivashchenko, A.; Rakhmetullina, A.; Aisina, D. How miRNAs can Protect Humans from Coronaviruses COVID-19, SARS-CoV, and MERS-CoV. Research Square. 2020. Available online: https://assets.researchsquare.com/files/rs-16264/v1/manuscript.pdf (accessed on 18 February 2021).
- Abplanalp, W.T.; Fischer, A.; John, D.; Zeiher, A.M.; Gosgnach, W.; Darville, H.; Montgomery, R.; Pestano, L.; Allée, G.; Paty, I.; et al. Efficiency and Target Derepression of An-ti-miR-92a: Results of a First in Human Study. Nucleic Acid Ther. 2020, 30, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2010, 39, D152–D157. [Google Scholar] [CrossRef] [Green Version]
- The RNAcentral Consortium. RNAcentral: A hub of information for non-coding RNA sequences. Nucleic Acids Res. 2019, 47, D221–D229. [Google Scholar] [CrossRef] [Green Version]
- Mann, M.; Wright, P.R.; Backofen, R. IntaRNA 2.0: Enhanced and customizable prediction of RNA–RNA interactions. Nucleic Acids Res. 2017, 45, W435–W439. [Google Scholar] [CrossRef]
- Lorenz, R.; Bernhart, S.H.F.; Zu Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26. [Google Scholar] [CrossRef]
- Umu, S.U.; Gardner, P.P. A comprehensive benchmark of RNA-RNA interaction prediction tools for all domains of life. Bioinformatics 2017, 33, 988–996. [Google Scholar] [CrossRef] [Green Version]
- Lai, D.; Meyer, I.M. A comprehensive comparison of general RNA–RNA interaction prediction methods. Nucleic Acids Res. 2015, 44, e61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, w202–w208. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanoria, S.; Rennie, W.; Liu, C.; Carmack, C.S.; Lu, J.; Ding, Y. STarMir Tools for Prediction of microRNA binding sites. Methods Mol. Biol. 2016, 1490, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Rennie, W.; Liu, C.; Carmack, C.S.; Wolenc, A.; Kanoria, S.; Lu, J.; Long, D.; Ding, Y. STarMir: A web server for prediction of microRNA binding sites. Nucleic Acids Res. 2014, 42, W114–W118. [Google Scholar] [CrossRef] [Green Version]
- Gruber, A.R.; Lorenz, R.; Bernhart, S.H.F.; Neuböck, R.; Hofacker, I.L. The Vienna RNA Websuite. Nucleic Acids Res. 2008, 36, W70–W74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartel, D.P. MicroRNAs: Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Xu, Y.; Xie, X.; Wang, T.; Ko, J.-H.; Zhou, T. The role of RNA structure at 5’ untranslated region in microRNA-mediated gene regulation. RNA 2014, 20, 1369–1375. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5’-UTR and 3’-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef] [Green Version]
- Santovito, D.; Egea, V.; Bidzhekov, K.; Natarelli, L.; Mourão, A.; Blanchet, X.; Wichapong, K.; Aslani, M.; Brunßen, C.; Horckmans, M.; et al. Noncanonical inhibition of caspase-3 by a nuclear microRNA confers endothelial protection by autophagy in atherosclerosis. Sci. Transl. Med. 2020, 12, eaaz2294. [Google Scholar] [CrossRef] [PubMed]
- Natarelli, L.; Geißler, C.; Csaba, G.; Wei, Y.; Zhu, M.; Di Francesco, A.; Hartmann, P.; Zimmer, R.; Schober, A. miR-103 promotes endothelial maladaptation by targeting lncWDR59. Nat. Commun. 2018, 9, 1–15. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, X. miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wang, X. Prediction of functional microRNA targets by integrative modeling of microRNA binding and target expression data. Genome Biol. 2019, 20, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Melo, D.; Nilsson-Payant, B.E.; Liu, W.C.; Uhl, S.; Hoagland, D.; Møller, R.; Jordan, T.X.; Oishi, K.; Panis, M.; Sachs, D.; et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell 2020, 181, 1036–1045. [Google Scholar] [CrossRef] [PubMed]
- Islam, A.; Khan, A.-A.-K. Lung biopsy cells transcriptional landscape from COVID-19 patient stratified lung injury in SARS-CoV-2 infection through impaired pulmonary surfactant metabolism. bioRxiv 2020. 2020.05.07.082297. Available online: https://www.biorxiv.org/content/10.1101/2020.05.07.082297v1 (accessed on 18 February 2021).
- Khongnomnan, K.; Makkoch, J.; Poomipak, W.; Poovorawan, Y.; Payungporn, S. Human miR-3145 inhibits influenza A viruses repli-cation by targeting and silencing viral PB1 gene. Exp. Biol. Med. 2015, 240, 1630–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.J.; Yang, J.; Fan, X.L.; Zhao, H.B.; Hu, W.; Li, Z.P.; Yu, G.C.; Ding, X.R.; Wang, J.Z.; Bo, X.C.; et al. Cellular microRNA let-7c inhibits M1 protein expression of the H1N1 influ-enza A virus in infected human lung epithelial cells. J. Cell. Mol. Med. 2012, 16, 2539–2546. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Martinez, R.; Farr, J.N.; Weivoda, M.M.; Negley, B.A.; Onken, J.L.; Thicke, B.S.; Fulcer, M.M.; Fraser, D.G.; van Wijnen, A.J.; Khosla, S.; et al. miR-219a-5p Regulates Rorβ During Osteo-blast Differentiation and in Age-related Bone Loss. J. Bone Miner. Res. 2019, 34, 135–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Zhai, P.; Ye, Z.; Deng, P.; Fan, Y.; Zeng, Y.; Pang, Z.; Zeng, J.; Li, J.; Feng, W. Differential expression of miR-195-5p in collapse of steroid-induced osteonecrosis of the femoral head. Oncotarget 2017, 8, 42638–42647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, R.; Ji, X.; Rong, R.; Li, Y.; Yao, W.; Yuan, J.; Wu, Q.; Yang, J.; Yan, W.; Han, L.; et al. MiR-449a regulates autophagy to inhibit silica-induced pulmonary fibrosis through targeting Bcl2. J. Mol. Med. 2016, 94, 1267–1279. [Google Scholar] [CrossRef]
- Zhang, X.; Zhao, X.; Sun, H.; Yan, Y.; Huang, L.; Gu, W.; Jiang, W.; Wang, Y.; Zhu, C.; Ji, W.; et al. The role of miR-29c/B7-H3 axis in children with allergic asthma. J. Transl. Med. 2018, 16, 1–10. [Google Scholar] [CrossRef]
- Courboulin, A.; Paulin, R.; Giguère, N.J.; Saksouk, N.; Perreault, T.; Meloche, J.; Paquet, E.R.; Biardel, S.; Provencher, S.; Côté, J.; et al. Role for miR-204 in human pulmonary arterial hypertension. J. Exp. Med. 2011, 208, 535–548. [Google Scholar] [CrossRef]
- Gupta, R.; Radicioni, G.; Abdelwahab, S.; Dang, H.; Carpenter, J.; Chua, M.; Mieczkowski, P.A.; Sheridan, J.T.; Randell, S.H.; Kesimer, M. Intercellular Communication between Airway Epithe-lial Cells Is Mediated by Exosome-Like Vesicles. Am. J. Respir. Cell Mol. Biol. 2019, 60, 209–220. [Google Scholar] [CrossRef] [PubMed]
- Latsoudis, H.; Mashreghi, M.F.; Grün, J.R.; Chang, H.D.; Stuhlmüller, B.; Repa, A.; Gergiannaki, I.; Kabouraki, E.; Vlachos, G.S.; Häupl, T.; et al. Differential Expression of miR-4520a Associated with Pyrin Mutations Suggesting a Role of Autophagy in Familial Mediterranean Fever (FMF). J. Cell. Physiol. 2017, 232, 1326–1336. [Google Scholar] [CrossRef]
- Chow, J.T.-S.; Salmena, L. Prediction and Analysis of SARS-CoV-2-Targeting MicroRNA in Human Lung Epithelium. Genes 2020, 11, 1002. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.; Daulatabad, S.V.; Srivastava, M.; Janga, S.C. Role of SARS-CoV-2 in Altering the RNA-Binding Protein and miRNA-Directed Post-Transcriptional Regulatory Networks in Humans. Int. J. Mol. Sci. 2020, 21, 7090. [Google Scholar] [CrossRef]
- He, L.; Yuan, J.; Xu, Q.; Chen, R.; Chen, L.; Fang, M. miRNA-1283 Regulates the PERK/ATF4 Pathway in Vascular Injury by Targeting ATF4. PLoS ONE 2016, 11, e0159171. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Bai, P.; Chen, Y.; Yu, T.; Li, F. Inhibition of miR-495 Improves Both Vascular Remodeling and Angiogenesis in Pulmonary Hypertension. J. Vasc. Res. 2019, 56, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Au, K.Y.; Pong, J.C.H.; Ling, W.L.; Li, J.C.B. MiR-1303 Regulates Mycobacteria Induced Autophagy by Targeting Atg2B. PLoS ONE 2016, 11, e0146770. [Google Scholar] [CrossRef] [Green Version]
- Ioannidis, J.; Donadeu, F.X. Comprehensive analysis of blood cells and plasma identifies tissue-specific miRNAs as potential novel circulating biomarkers in cattle. BMC Genom. 2018, 19, 243. [Google Scholar] [CrossRef] [Green Version]
- Stolzenburg, L.R.; Wachtel, S.; Dang, H.; Harris, A. miR-1343 attenuates pathways of fibrosis by targeting the TGF-β receptors. Biochem. J. 2016, 473, 245–256. [Google Scholar] [CrossRef] [Green Version]
- Stolzenburg, L.R.; Harris, A. Microvesicle-mediated delivery of miR-1343: Impact on markers of fibrosis. Cell Tissue Res. 2017, 371, 325–338. [Google Scholar] [CrossRef]
- Rothschild, S.I.; Tschan, M.P.; Jaggi, R.; Fey, M.F.; Gugger, M.; Gautschi, O. MicroRNA-381 represses ID1 and is deregulated in lung ade-nocarcinoma. J. Thorac. Oncol. 2012, 7, 1069–1077. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, L.; Zhu, H.; Zhang, L.; Wang, R.; Zhang, Z.; Huang, J.; Zhang, S.; Jian, F.; Ning, C.; et al. MicroRNA expression profile of HCT-8 cells in the early phase of Cryptosporidium parvum infection. BMC Genom. 2019, 20, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L; Yu, S. Role of miR-520b in non-small cell lung cancer. Exp. Ther. Med. 2018, 16, 3987–3995. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.; Cai, Q.; Zhang, X.; Zhang, H.; Zhong, Y.; Xu, C.; Li, Y. Two less common human microRNAs miR-875 and miR-3144 target a conserved site of E6 oncogene in most high-risk human papillomavirus subtypes. Protein Cell 2015, 6, 575–588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Yang, Y.-E.; Jin, J.; Zhang, M.-Y.; Liu, X.-X.; Yin, Y.-H.; Qu, Y.-Q. Identification of lncRNA biomarkers in lung squamous cell carcinoma using comprehensive analysis of lncRNA mediated ceRNA network. Artif. Cells Nanomedicine Biotechnol. 2019, 47, 3246–3258. [Google Scholar] [CrossRef] [Green Version]
- Biju, B.M.; Chhabra, S.S.; Sarkar, C. Elucidation of novel miRNA candidates and their role in unraveling the pathology of Non-Alcoholic Fatty Liver Disease. bixRxiv 2020. Available online: https://www.biorxiv.org/content/biorxiv/early/2020/02/02/2020.01.31.917831.full.pdf (accessed on 18 February 2021).
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135. [Google Scholar] [CrossRef]
- Wu, H.; Liu, H.Y.; Liu, W.J.; Shi, Y.L.; Bao, D. miR-377-5p inhibits lung cancer cell proliferation, invasion, and cell cycle progression by targeting AKT1 signaling. J. Cell. Biochem. 2019, 120, 8120–8128. [Google Scholar] [CrossRef]
- Fan, J.; An, X.; Yang, Y.; Xu, H.; Fan, L.; Deng, L.; Linyuan, F.; Weng, X.; Zhang, J.; Zhao, R.C. MiR-1292 Targets FZD4 to Regulate Senescence and Osteogenic Differentiation of Stem Cells in TE/SJ/Mesenchymal Tissue System via the Wnt/β-catenin Pathway. Aging Dis. 2018, 9, 1103–1121. [Google Scholar] [CrossRef] [Green Version]
- Mao, L.; Liu, S.; Hu, L.; Jia, L.; Wang, H.; Guo, M.; Chen, C.; Liu, Y.; Xu, L. miR-30 Family: A Promising Regulator in Development and Disease. BioMed Res. Int. 2018, 2018, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Huang, C.; Yang, Q.; Gao, L.; Liu, H.-C.; Tang, J.; Feng, W.-H. MicroRNA-30c Modulates Type I IFN Responses To Facilitate Porcine Reproductive and Respiratory Syndrome Virus Infection by Targeting JAK1. J. Immunol. 2016, 196, 2272–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, R.; Wang, Y.; Wan, J.; Leng, D.; Gong, J.; Li, J.; Liang, Y.; Zhai, Z.; Yang, Y. Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine 2017, 96, e7354. [Google Scholar] [CrossRef]
- Keller, A.; Ludwig, N.; Fehlmann, T.; Kahraman, M.; Backes, C.; Kern, F.; Vogelmeier, C.F.; Diener, C.; Fischer, U.; Biertz, F.; et al. Low miR-150-5p and miR-320b Expression Predicts Reduced Survival of COPD Patients. Cells 2019, 8, 1162. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Wang, J.; Su, H. MiR-3150b inhibits hepatocellular carcinoma cell proliferation, migration and invasion by targeting GOLPH3. J. Investig. Med. 2019, 68, 770–775. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.N.; Luo, S.; Liu, C.; Piao, Z.; Gou, W.; Wang, Y.; Guan, W.; Li, Q.; Zou, H.; Yang, Z.Z.; et al. miR-491 Inhibits Osteosarcoma Lung Metastasis and Chemoresistance by Targeting αB-crystallin. Mol. Ther. 2017, 25, 2140–2149. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Lin, J.; Jin, Y.; Chen, M.; Zheng, H.; Feng, J. The miRNA hsa-miR-6515-3p potentially contributes to lncRNA H19-mediated-lung cancer metastasis. J. Cell. Biochem. 2019, 120, 17413–17421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.H.; Long, L.; Luo, Z.X.; Li, L.M.; You, J.R. Anti-inflammatory role of microRNA let-7c in LPS treated alveolar macrophages by tar-geting STAT3. Asian Pac. J. Trop. Med. 2016, 9, 72–75. [Google Scholar] [CrossRef] [Green Version]
- Higaki, M.; Shintani, T.; Hamada, A.; Rosli, S.N.Z.; Okamoto, T. Eldecalcitol (ED-71)-induced exosomal miR-6887-5p suppresses squa-mous cell carcinoma cell growth by targeting heparin-binding protein 17/fibroblast growth factor–binding protein-1 (HBp17/FGFBP-1). In Vitro Cell. Dev. Biol. Anim. 2020, 56, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Chooniedass-Kothari, S.; Emberley, E.; Hamedani, M.K.; Troup, S.; Wang, X.; Czosnek, A.; Hube, F.; Mutawe, M.; Watson, P.H.; Leygue, E. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett. 2004, 566, 43–47. [Google Scholar] [CrossRef]
- Yao, R.-W.; Wang, Y.; Chen, L.-L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551. [Google Scholar] [CrossRef]
- Somarowthu, S.; Legiewicz, M.; Chillón, I.; Marcia, M.; Liu, F.; Pyle, A.M. HOTAIR Forms an Intricate and Modular Secondary Structure. Mol. Cell 2015, 58, 353–361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Su, H.; Xu, X.; Yan, C.; Shi, Y.; Hu, Y.; Dong, L.; Ying, S.; Ying, K.; Zhang, R. LncRNA H19 promotes the proliferation of pulmonary artery smooth muscle cells through AT1R via sponging let-7b in monocrotaline-induced pulmonary arterial hypertension. Respir. Res. 2018, 19, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteil, V.; Kwon, H.; Prado, P.; Hagelkrüys, A.; Wimmer, R.A.; Stahl, M.; Leopoldi, A.; Garreta, E.; Del Pozo, C.H.; Prosper, F.; et al. Inhibition of SARS-CoV-2 Infections in Engineered Human Tissues Using Clinical-Grade Soluble Human ACE2. Cell 2020, 181, 905–913. [Google Scholar] [CrossRef] [PubMed]
- Imai, Y.; Kuba, K.; Rao, S.; Huan, Y.; Guo, F.; Guan, B.; Yang, P.; Sarao, R.; Wada, T.; Leong-Poi, H.; et al. Angiotensin-converting enzyme 2 protects from severe acute lung failure. Nat. Cell Biol. 2005, 436, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Rolle, K.; Piwecka, M.; Belter, A.; Wawrzyniak, D.; Jeleniewicz, J.; Barciszewska, M.Z.; Barciszewski, J. The Sequence and Structure Determine the Function of Mature Human miRNAs. PLoS ONE 2016, 11, e0151246. [Google Scholar] [CrossRef] [PubMed]
- Krützfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689. [Google Scholar] [CrossRef]
- Lucas, T.; Bonauer, A.; Dimmeler, S. RNA Therapeutics in Cardiovascular Disease. Circ. Res. 2018, 123, 205–220. [Google Scholar] [CrossRef]
- van Rooij, E.; Kauppinen, S. Development of microRNA therapeutics is coming of age. EMBO Mol. Med. 2014, 6, 851–864. [Google Scholar] [CrossRef]
- Fanciullino, R.; Ciccolini, J.; Milano, G. COVID-19 vaccine race: Watch your step for cancer patients. Br. J. Cancer 2020, 1–2. [Google Scholar] [CrossRef]
- Janssen, H.L.; Reesink, H.W.; Lawitz, E.J.; Zeuzem, S.; Rodriguez-Torres, M.; Patel, K.; Van Der Meer, A.J.; Patick, A.K.; Chen, A.; Zhou, Y.; et al. Treatment of HCV infection by targeting mi-croRNA. N. Engl. J. Med. 2013, 368, 1685–1694. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.-K.; Kafert-Kasting, S.; Thum, T. Preclinical and Clinical Development of Noncoding RNA Therapeutics for Cardiovascular Disease. Circ. Res. 2020, 126, 663–678. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Li, W.; Drabek, D.; Okba, N.M.; van Haperen, R.; Osterhaus, A.D.; van Kuppeveld, F.J.; Haagmans, B.L.; Grosveld, F.; Bosch, B.J. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat. Commun. 2020, 11, 2251. [Google Scholar] [CrossRef] [PubMed]





| miRNAs | Reported Function | Reference |
|---|---|---|
| hsa-miR-1283 | Endothelial vascular injury | [46] |
| hsa-miR-495-5p | Inhibits vascular remodeling and angiogenesis in PAH | [47] |
| hsa-miR-1303 | Regulate the autophagy process in mycobacteria infection | [48] |
| hsa-miR-204-3p | Promoter of PAH | [41] |
| hsa-miR-6529-5p | Novel potential tissue specific biomarker in cattle | [49] |
| hsa-miR-1343-3p | Attenuate fibrosis in fibrotic lung disease/microvesicle | [50,51] |
| hsa-miR-3661 | Direct involvement with SARS-CoV-2 proteins from lung biopsy | [34] |
| hsa-miR-381-3p | Deregulated in lung adenocarcinoma | [52] |
| hsa-miR-3976 | Regulates apoptosis in hosts after microbial infection | [53] |
| hsa-miR-520b-5p | Inhibits NSCLC | [54] |
| hsa-miR-3144-5p | Interact with viral proteins | [55] |
| hsa-miR-4652-5p | Lung cancer expressed miRNA | [56] |
| hsa-miR-6857-5p | Prognostic of viral-related cervical cancer/marker of NSCLC | [57] |
| hsa-miR-377-5p | Promote fibronectin production/inhibits lung cell proliferation | [58,59] |
| hsa-miR-1292-5p | Inhibitor of osteogenic differentiation, promotes osteoporosis | [60] |
| hsa-miR-219a | Arthritis/NSCLC | [37] |
| hsa-miR-30c-1-3p | Positive bone development/promotes viral infection | [61,62] |
| hsa-miR-449a | Inhibits pulmonary fibrosis | [39] |
| hsa-miR-5572 | Upregulated in osteonecrosis femoral head | [38] |
| hsa-miR-6752-5p | Highly expressed in airway epithelial cells/mucin overproduction | [42] |
| hsa-miR-4531 | Upregulated in children with asthma | [40] |
| hsa-miR-6831-3p | Anti-atherogenic/PAH | [42,63] |
| hsa-miR-377-5p | Inhibits lung cancer cell proliferation | [59] |
| hsa-miR-3123 | Negative correlation with survival of COPD patients | [64] |
| hsa-miR-3150b-3p | Inhibits cell proliferation in NSCLC patients | [65] |
| hsa-miR-451b | Inhibits osteosarcoma lung metastasis | [66] |
| hsa-miR-4520-3p | Associated with FMF-related mutations | [43] |
| hsa-miR-491-5p | Inhibits osteosarcoma lung metastasis | [66] |
| hsa-miR-6515 | Contributes to lncRNA H19-mediated lung cancer metastasis | [67] |
| hsa-let-7c-5p | Inhibits H1N1 protein synthesis/anti-inflammatory role in COPD | [33,36,68] |
| hsa-miR-6887-5p | Inhibits squamous cell carcinoma cell growth | [69] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Natarelli, L.; Parca, L.; Mazza, T.; Weber, C.; Virgili, F.; Fratantonio, D. MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2. Non-Coding RNA 2021, 7, 14. https://doi.org/10.3390/ncrna7010014
Natarelli L, Parca L, Mazza T, Weber C, Virgili F, Fratantonio D. MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2. Non-Coding RNA. 2021; 7(1):14. https://doi.org/10.3390/ncrna7010014
Chicago/Turabian StyleNatarelli, Lucia, Luca Parca, Tommaso Mazza, Christian Weber, Fabio Virgili, and Deborah Fratantonio. 2021. "MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2" Non-Coding RNA 7, no. 1: 14. https://doi.org/10.3390/ncrna7010014
APA StyleNatarelli, L., Parca, L., Mazza, T., Weber, C., Virgili, F., & Fratantonio, D. (2021). MicroRNAs and Long Non-Coding RNAs as Potential Candidates to Target Specific Motifs of SARS-CoV-2. Non-Coding RNA, 7(1), 14. https://doi.org/10.3390/ncrna7010014