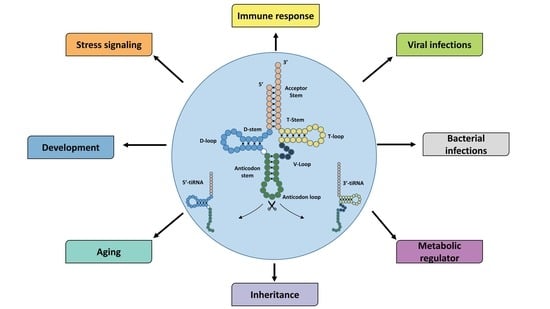
tiRNAs: Insights into Their Biogenesis, Functions, and Future Applications in Livestock Research
Abstract
:1. Introduction
2. tiRNA Classification, Biogenesis, and Subcellular Localisation
3. Regulation of tiRNA Processing by Angiogenin
4. Role of tiRNA in Development, Cell Differentiation, and Apoptosis
5. Role of tiRNA on Immunity and Their Potential as Cross-Species Biomarkers
6. Role of tiRNAs in Stress Signalling and Behaviour
7. tsRNA Databases
8. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vavouri, T.; Walter, K.; Gilks, W.R.; Lehner, B.; Elgar, G. Parallel Evolution of Conserved Non-Coding Elements That Target a Common Set of Developmental Regulatory Genes from Worms to Humans. Genome Biol. 2007, 8, R15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siepel, A.; Bejerano, G.; Pedersen, J.S.; Hinrichs, A.S.; Hou, M.; Rosenbloom, K.; Clawson, H.; Spieth, J.; Hillier, L.W.; Richards, S.; et al. Evolutionarily Conserved Elements in Vertebrate, Insect, Worm, and Yeast Genomes. Genome Res. 2005, 15, 1034–1050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.; Skogerbø, G.; Chen, R. Conserved Distances between Vertebrate Highly Conserved Elements. Hum. Mol. Genet. 2006, 15, 2911–2922. [Google Scholar] [CrossRef] [Green Version]
- Kapusta, A.; Feschotte, C. Volatile Evolution of Long Noncoding RNA Repertoires: Mechanisms and Biological Implications. Trends Genet. 2014, 30, 439–452. [Google Scholar] [CrossRef] [Green Version]
- Mattick, J.S. Non-Coding RNAs: The Architects of Eukaryotic Complexity. EMBO Rep. 2001, 2, 986–991. [Google Scholar] [CrossRef]
- Sana, J.; Faltejskova, P.; Svoboda, M.; Slaby, O. Novel Classes of Non-Coding RNAs and Cancer. J. Transl. Med. 2012, 10, 103. [Google Scholar] [CrossRef] [Green Version]
- Sun, Y.-M.; Chen, Y.-Q. Principles and Innovative Technologies for Decrypting Noncoding RNAs: From Discovery and Functional Prediction to Clinical Application. J. Hematol. Oncol. 2020, 13, 109. [Google Scholar] [CrossRef]
- Wang, J.; Samuels, D.C.; Zhao, S.; Xiang, Y.; Zhao, Y.Y.; Guo, Y. Current Research on Non-Coding Ribonucleic Acid (RNA). Genes 2017, 8, 366. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Dana, H.; Chalbatani, G.M.; Mahmoodzadeh, H.; Karimloo, R.; Rezaiean, O.; Moradzadeh, A.; Mehmandoost, N.; Moazzen, F.; Mazraeh, A.; Marmari, V.; et al. Molecular Mechanisms and Biological Functions of SiRNA. Int. J. Biomed. Sci. 2017, 13, 48–57. [Google Scholar]
- Lim, R.S.M.; Kai, T. A Piece of the Pi(e): The Diverse Roles of Animal PiRNAs and Their PIWI Partners. Semin. Cell Dev. Biol. 2015, 47–48, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Wen, J.; Huang, Z.; Chen, X.P.; Zhang, B.X.; Chu, L. Small Nucleolar RNAs: Insight into Their Function in Cancer. Front. Oncol. 2019, 9, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindsay, J.; Carone, D.M.; Brown, J.; Hall, L.; Qureshi, S.; Mitchell, S.E.; Jannetty, N.; Hannon, G.; Renfree, M.; Pask, A.; et al. Unique Small RNA Signatures Uncovered in the Tammar Wallaby Genome. BMC Genom. 2012, 13, 559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, F.; Li, X.; Hiew, S.; Brady, H.; Liu, Y.; Dou, Y. Dicer Independent Small RNAs Associate with Telomeric Heterochromatin. RNA 2009, 15, 1274–1281. [Google Scholar] [CrossRef] [Green Version]
- Evangelista, A.F.; de Menezes, W.P.; Berardinelli, G.N.; dos Santos, W.; Scapulatempo-Neto, C.; Guimarães, D.P.; Calin, G.A.; Reis, R.M. Pyknon-Containing Transcripts Are Downregulated in Colorectal Cancer Tumors, and Loss of PYK44 Is Associated with Worse Patient Outcome. Front. Genet. 2020, 11, 1212. [Google Scholar] [CrossRef]
- Zhu, L.; Ge, J.; Li, T.; Shen, Y.; Guo, J. TRNA-Derived Fragments and TRNA Halves: The New Players in Cancers. Cancer Lett. 2019, 452, 31–37. [Google Scholar] [CrossRef]
- Tao, E.W.; Cheng, W.Y.; Li, W.L.; Yu, J.; Gao, Q.Y. TiRNAs: A Novel Class of Small Noncoding RNAs That Helps Cells Respond to Stressors and Plays Roles in Cancer Progression. J. Cell. Physiol. 2020, 235, 683–690. [Google Scholar] [CrossRef]
- Liu, B.; Cao, J.; Wang, X.; Guo, C.; Liu, Y.; Wang, T. Deciphering the TRNA-Derived Small RNAs: Origin, Development, and Future. Cell Death Dis. 2021, 13, 24. [Google Scholar] [CrossRef]
- Su, Z.; Frost, E.L.; Lammert, C.R.; Przanowska, R.K.; Lukens, J.R.; Dutta, A. TRNA-Derived Fragments and MicroRNAs in the Maternal-Fetal Interface of a Mouse Maternal-Immune-Activation Autism Model. RNA Biol. 2020, 17, 1183–1195. [Google Scholar] [CrossRef]
- Maute, R.L.; Schneider, C.; Sumazin, P.; Holmes, A.; Califano, A.; Basso, K.; Dalla-Favera, R. TRNA-Derived MicroRNA Modulates Proliferation and the DNA Damage Response and Is down-Regulated in B Cell Lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 1404–1409. [Google Scholar] [CrossRef] [Green Version]
- Cammas, A.; Millevoi, S. RNA G-Quadruplexes: Emerging Mechanisms in Disease. Nucleic Acids Res. 2017, 45, 1584–1595. [Google Scholar] [CrossRef] [Green Version]
- Gao, Z.W.; Wang, X.; Zhang, H.Z.; Lin, F.; Liu, C.; Dong, K. The Roles of Adenosine Deaminase in Autoimmune Diseases. Autoimmun. Rev. 2021, 20, 102709. [Google Scholar] [CrossRef]
- Xu, H.; Chen, W.; Zheng, F.; Tang, D.; Dai, W.; Huang, S.; Zhang, C.; Zeng, J.; Wang, G.; Dai, Y. The Potential Role of TRNAs and Small RNAs Derived from TRNAs in the Occurrence and Development of Systemic Lupus Erythematosus. Biochem. Biophys. Res. Commun. 2020, 527, 561–567. [Google Scholar] [CrossRef]
- Hopper, A.K.; Nostramo, R.T. TRNA Processing and Subcellular Trafficking Proteins Multitask in Pathways for Other RNAs. Front. Genet. 2019, 10, 96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krahn, N.; Fischer, J.T.; Söll, D. Naturally Occurring TRNAs with Non-Canonical Structures. Front. Microbiol. 2020, 11, 2616. [Google Scholar] [CrossRef]
- Phizicky, E.M.; Hopper, A.K. TRNA Biology Charges to the Front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [Green Version]
- Yu, M.; Lu, B.; Zhang, J.; Ding, J.; Liu, P.; Lu, Y. TRNA-Derived RNA Fragments in Cancer: Current Status and Future Perspectives. J. Hematol. Oncol. 2020, 13, 121. [Google Scholar] [CrossRef]
- Cao, J.; Cowan, D.B.; Wang, D.-Z. TRNA-Derived Small RNAs and Their Potential Roles in Cardiac Hypertrophy. Front. Pharm. 2020, 11, 572941. [Google Scholar] [CrossRef]
- Yue, T.; Zhan, X.; Zhang, D.; Jain, R.; Wang, K.W.; Choi, J.H.; Misawa, T.; Su, L.; Quan, J.; Hildebrand, S.; et al. SLFN2 Protection of TRNAs from Stress-Induced Cleavage Is Essential for T Cell–Mediated Immunity. Science 2021, 372, eaba4220. [Google Scholar] [CrossRef]
- Telonis, A.G.; Loher, P.; Honda, S.; Jing, Y.; Palazzo, J.; Kirino, Y.; Rigoutsos, I. Dissecting TRNA-Derived Fragment Complexities Using Personalized Transcriptomes Reveals Novel Fragment Classes and Unexpected Dependencies. Oncotarget 2015, 6, 24797–24822. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Xu, W.; Liu, K.; Jiang, Z.; Han, Y.; Jin, H.; Zhang, L.; Shen, W.; Jia, S.; Sun, Q.; et al. 5′ Half of Specific TRNAs Feeds Back to Promote Corresponding TRNA Gene Transcription in Vertebrate Embryos. Sci. Adv. 2021, 7, eabh0494. [Google Scholar] [CrossRef]
- Blanco, S.; Dietmann, S.; Flores, J.V.; Hussain, S.; Kutter, C.; Humphreys, P.; Lukk, M.; Lombard, P.; Treps, L.; Popis, M.; et al. Aberrant Methylation of t RNA s Links Cellular Stress to Neuro-developmental Disorders. EMBO J. 2014, 33, 2020–2039. [Google Scholar] [CrossRef]
- Schaefer, M.; Pollex, T.; Hanna, K.; Tuorto, F.; Meusburger, M.; Helm, M.; Lyko, F. RNA Methylation by Dnmt2 Protects Transfer RNAs against Stress-Induced Cleavage. Genes Dev. 2010, 24, 1590–1595. [Google Scholar] [CrossRef] [Green Version]
- Levitz, R.; Chapman, D.; Amitsur, M.; Green, R.; Snyder, L.; Kaufmann, G. The Optional E. coli Prr Locus Encodes a Latent Form of Phage T4-Induced Anticodon Nuclease. EMBO J. 1990, 9, 1383–1389. [Google Scholar] [CrossRef]
- Zhao, H.; Bojanowski, K.; Ingber, D.E.; Panigrahy, D.; Pepper, M.S.; Montesano, R.; Shing, Y. New Role for TRNA and Its Fragment Purified from Human Urinary Bladder Carcinoma Conditioned Medium: Inhibition of Endothelial Cell Growth. J. Cell. Biochem. 2000, 76, 109–117. [Google Scholar] [CrossRef]
- Yamasaki, S.; Ivanov, P.; Hu, G.F.; Anderson, P. Angiogenin Cleaves TRNA and Promotes Stress-Induced Translational Repression. J. Cell Biol. 2009, 185, 35–42. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Feng, J.; Liu, Q.; Sun, F.; Tie, Y.; Zhu, J.; Xing, R.; Sun, Z.; Zheng, X. Stress Induces TRNA Cleavage by Angiogenin in Mammalian Cells. FEBS Lett. 2009, 583, 437–442. [Google Scholar] [CrossRef] [Green Version]
- Sobala, A.; Hutvagner, G. Transfer RNA-Derived Fragments: Origins, Processing, and Functions. Wiley Interdiscip. Rev. RNA 2011, 2, 853–862. [Google Scholar] [CrossRef] [Green Version]
- Prehn, J.H.M.; Jirström, E. Angiogenin and TRNA Fragments in Parkinson’s Disease and Neurodegeneration. Acta Pharmacol. Sin. 2020, 41, 442–446. [Google Scholar] [CrossRef]
- Zhao, F.; Cheng, L.; Shao, Q.; Chen, Z.; Lv, X.; Li, J.; He, L.; Sun, Y.; Ji, Q.; Lu, P.; et al. Characterization of Serum Small Extracellular Vesicles and Their Small RNA Contents across Humans, Rats, and Mice. Sci. Rep. 2020, 10, 4197. [Google Scholar] [CrossRef]
- Salinas-Giegé, T.; Giegé, R.; Giegé, P. TRNA Biology in Mitochondria. Int. J. Mol. Sci. 2015, 16, 4518–4559. [Google Scholar] [CrossRef] [Green Version]
- Telonis, A.G.; Kirino, Y.; Rigoutsos, I. Mitochondrial TRNA-Lookalikes in Nuclear Chromosomes: Could They Be Functional? RNA Biol. 2015, 12, 375–380. [Google Scholar] [CrossRef] [Green Version]
- Meseguer, S. MicroRNAs and TRNA-Derived Small Fragments: Key Messengers in Nuclear–Mitochondrial Communication. Front. Mol. Biosci. 2021, 8, 203. [Google Scholar] [CrossRef]
- Maniataki, E.; Mourelatos, Z. Human Mitochondrial TRNAMet Is Exported to the Cytoplasm and Associates with the Argonaute 2 Protein. RNA 2005, 11, 849–852. [Google Scholar] [CrossRef] [Green Version]
- Rubio, M.A.T.; Rinehart, J.J.; Krett, B.; Duvezin-Caubet, S.; Reichert, A.S.; Söll, D.; Alfonzo, J.D. Mammalian Mitochondria Have the Innate Ability to Import TRNAs by a Mechanism Distinct from Protein Import. Proc. Natl. Acad. Sci. USA 2008, 105, 9186–9191. [Google Scholar] [CrossRef] [Green Version]
- Fett, J.W.; Strydom, D.J.; Lobb, R.R.; Alderman, E.M.; Bethune, J.L.; Riordan, J.F.; Vallee, B.L. Isolation and Characterization of Angiogenin, an Angiogenic Protein from Human Carcinoma Cells. Biochemistry 1985, 24, 5480–5486. [Google Scholar] [CrossRef]
- Li, S.; Ibaragi, S.; Hu, G.-F. Angiogenin as a Molecular Target for the Treatment of Prostate Cancer. Curr. Cancer Ther. Rev. 2012, 7, 83–90. [Google Scholar] [CrossRef] [Green Version]
- Saxena, S.K.; Rybak, S.M.; Davey, R.T.; Youle, R.J.; Ackerman, E.J. Angiogenin Is a Cytotoxic, TRNA-Specific Ribonuclease in the RNase A Superfamily. J. Biol. Chem. 1992, 267, 21982–21986. [Google Scholar] [CrossRef]
- Cho, S.; Beintema, J.J.; Zhang, J. The Ribonuclease A Superfamily of Mammals and Birds: Identifying New Members and Tracing Evolutionary Histories. Genomics 2005, 85, 208–220. [Google Scholar] [CrossRef]
- Nitto, T.; Dyer, K.D.; Czapiga, M.; Rosenberg, H.F. Evolution and Function of Leukocyte RNase A Ribonucleases of the Avian Species, Gallus Gallus. J. Biol. Chem. 2006, 281, 25622–25634. [Google Scholar] [CrossRef] [Green Version]
- Sheng, J.; Xu, Z. Three Decades of Research on Angiogenin: A Review and Perspective. Acta Biochim. Biophys. Sin. 2016, 48, 399–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizzo, E.; Buonanno, P.; di Maro, A.; Ponticelli, S.; de Falco, S.; Quarto, N.; Cubellis, M.V.; D’Alessio, G. Ribonucleases and Angiogenins from Fish. J. Biol. Chem. 2006, 281, 27454–27460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, D.M.; Parker, R. The RNase Rny1p Cleaves TRNAs and Promotes Cell Death during Oxidative Stress in Saccharomyces Cerevisiae. J. Cell Biol. 2009, 185, 43–50. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, R.; Vallee, B.L. Human Placental Ribonuclease Inhibitor Abolishes Both Angiogenic and Ribonucleolytic Activities of Angiogenin. Proc. Natl. Acad. Sci. USA 1987, 84, 2238–2241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pizzo, E.; Sarcinelli, C.; Sheng, J.; Fusco, S.; Formiggini, F.; Netti, P.; Yu, W.; D’Alessio, G.; Hu, G.F. Ribonuclease/Angiogenin Inhibitor 1 Regulates Stressinduced Subcellular Localization of Angiogenin to Control Growth and Survival. J. Cell Sci. 2013, 126, 4308–4319. [Google Scholar] [CrossRef] [Green Version]
- Lai, K.; Luo, C.; Zhang, X.; Ye, P.; Zhang, Y.-D.; He, J.; Yao, K. Regulation of Angiogenin Expression and Epithelial-Mesenchymal Transition by HIF-1α Signaling in Hypoxic Retinal Pigment Epithelial Cells. Biochim. Biophys. Acta-Mol. Basis Dis. 2016, 1862, 1594–1607. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Kuscu, C.; Malik, A.; Shibata, E.; Dutta, A. Angiogenin Generates Specific Stress-Induced TRNA Halves and Is Not Involved in TRF-3–Mediated Gene Silencing. J. Biol. Chem. 2019, 294, 16930–16941. [Google Scholar] [CrossRef]
- Saikia, M.; Krokowski, D.; Guan, B.J.; Ivanov, P.; Parisien, M.; Hu, G.F.; Anderson, P.; Pan, T.; Hatzoglou, M. Genome-Wide Identification and Quantitative Analysis of Cleaved TRNA Fragments Induced by Cellular Stress. J. Biol. Chem. 2012, 287, 42708–42725. [Google Scholar] [CrossRef] [Green Version]
- Rybak, S.M.; Vallee, B.L. Base Cleavage Specificity of Angiogenin with Saccharomyces Cerevisiae and Escherichia Coli 5S RNAs. Biochemistry 1988, 27, 2288–2294. [Google Scholar] [CrossRef]
- Tosar, J.P.; Gámbaro, F.; Darré, L.; Pantano, S.; Westhof, E.; Cayota, A. Dimerization Confers Increased Stability to Nucleases in 5 Halves from Glycine and Glutamic Acid TRNAs. Nucleic Acids Res. 2018, 46, 9081–9093. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.; Wilson, B.; Kumar, P.; Dutta, A. Noncanonical Roles of TRNAs: TRNA Fragments and Beyond. Annu. Rev. Genet. 2020, 54, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Megel, C.; Hummel, G.; Lalande, S.; Ubrig, E.; Cognat, V.; Morelle, G.; Salinas-Giegé, T.; Duchêne, A.M.; Maréchal-Drouard, L. Plant RNases T2, but Not Dicer-like Proteins, Are Major Players of TRNA-Derived Fragments Biogenesis. Nucleic Acids Res. 2019, 47, 941–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, K.L.; Collins, K. Several RNase T2 Enzymes Function in Induced TRNA and RRNA Turnover in the Ciliate Tetrahymena. Mol. Biol. Cell 2012, 23, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Akiyama, Y.; Lyons, S.; Fay, M.M.; Abe, T.; Anderson, P.; Ivanov, P. Multiple Ribonuclease A Family Members Cleave Transfer RNAs in Response to Stress. bioRxiv 2019, 811174. [Google Scholar] [CrossRef]
- Donovan, J.; Rath, S.; Kolet-Mandrikov, D.; Korennykh, A. Rapid RNase L-Driven Arrest of Protein Synthesis in the DsRNA Response without Degradation of Translation Machinery. RNA 2017, 23, 1660–1671. [Google Scholar] [CrossRef] [Green Version]
- Nechooshtan, G.; Yunusov, D.; Chang, K.; Gingeras, T.R. Processing by RNase 1 Forms TRNA Halves and Distinct y RNA Fragments in the Extracellular Environment. Nucleic Acids Res. 2020, 48, 8035–8049. [Google Scholar] [CrossRef]
- Yang, J.Y.; Deng, X.Y.; Li, Y.S.; Ma, X.C.; Feng, J.X.; Yu, B.; Chen, Y.; Luo, Y.L.; Wang, X.; Chen, M.L.; et al. Structure of Schlafen13 Reveals a New Class of TRNA/RRNA- Targeting RNase Engaged in Translational Control. Nat. Commun. 2018, 9, 1165. [Google Scholar] [CrossRef] [Green Version]
- di Fazio, A.; Schlackow, M.; Pong, S.K.; Alagia, A.; Gullerova, M. Dicer Dependent TRNA Derived Small RNAs Promote Nascent RNA Silencing. Nucleic Acids Res. 2022, 50, 1734–1752. [Google Scholar] [CrossRef]
- Krishna, S.; Yim, D.G.; Lakshmanan, V.; Tirumalai, V.; Koh, J.L.; Park, J.E.; Cheong, J.K.; Low, J.L.; Lim, M.J.; Sze, S.K.; et al. Dynamic Expression of TRNA-derived Small RNAs Define Cellular States. EMBO Rep. 2019, 20, e47789. [Google Scholar] [CrossRef]
- Stefani, G.; Slack, F.J. Small Non-Coding RNAs in Animal Development. Nat. Rev. Mol. Cell Biol. 2008, 9, 219–230. [Google Scholar]
- Torres, A.G.; Reina, O.; Attolini, C.S.O.; de Pouplana, L.R. Differential Expression of Human TRNA Genes Drives the Abundance of TRNA-Derived Fragments. Proc. Natl. Acad. Sci. USA 2019, 116, 8451–8456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aoi, T.; Yae, K.; Nakagawa, M.; Ichisaka, T.; Okita, K.; Takahashi, K.; Chiba, T.; Yamanaka, S. Generation of Pluripotent Stem Cells from Adult Mouse Liver and Stomach Cells. Science 2008, 321, 699–702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.Y.; Ming, G.L.; Song, H. PUS7: A Targetable Epitranscriptomic Regulator of Glioblastoma Growth. Trends Pharmacol. Sci. 2021, 42, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Guzzi, N.; Cieśla, M.; Ngoc, P.C.T.; Lang, S.; Arora, S.; Dimitriou, M.; Pimková, K.; Sommarin, M.N.E.; Munita, R.; Lubas, M.; et al. Pseudouridylation of TRNA-Derived Fragments Steers Translational Control in Stem Cells. Cell 2018, 173, 1204–1216.e26. [Google Scholar] [CrossRef] [Green Version]
- Lyons, S.M.; Gudanis, D.; Coyne, S.M.; Gdaniec, Z.; Ivanov, P. Identification of Functional Tetramolecular RNA G-Quadruplexes Derived from Transfer RNAs. Nat. Commun. 2017, 8, 1127. [Google Scholar] [CrossRef] [Green Version]
- Lyons, S.M.; Kharel, P.; Akiyama, Y.; Ojha, S.; Dave, D.; Tsvetkov, V.; Merrick, W.; Ivanov, P.; Anderson, P. EIF4G Has Intrinsic G-Quadruplex Binding Activity That Is Required for TiRNA Function. Nucleic Acids Res 2020, 48, 6223–6233. [Google Scholar] [CrossRef]
- Yuan, Y.; Li, J.; He, Z.; Fan, X.; Mao, X.; Yang, M.; Yang, D. Trna-Derived Fragments as New Hallmarks of Aging and Age-Related Diseases. Aging Dis. 2021, 12, 1304–1322. [Google Scholar] [CrossRef]
- Victoria, B.; Dhahbi, J.M.; Nunez Lopez, Y.O.; Spinel, L.; Atamna, H.; Spindler, S.R.; Masternak, M.M. Circulating MicroRNA Signature of Genotype-by-Age Interactions in the Long-Lived Ames Dwarf Mouse. Aging Cell 2015, 14, 1055–1066. [Google Scholar] [CrossRef]
- Zhu, L.; Li, T.; Shen, Y.; Yu, X.; Xiao, B.; Guo, J. Using TRNA Halves as Novel Biomarkers for the Diagnosis of Gastric Cancer. Cancer Biomark. 2019, 25, 169–176. [Google Scholar] [CrossRef]
- Ivanov, P.; Emara, M.M.; Villen, J.; Gygi, S.P.; Anderson, P. Angiogenin-Induced TRNA Fragments Inhibit Translation Initiation. Mol. Cell 2011, 43, 613–623. [Google Scholar] [CrossRef] [Green Version]
- Mo, D.; Jiang, P.; Yang, Y.; Mao, X.; Tan, X.; Tang, X.; Wei, D.; Li, B.; Wang, X.; Tang, L.; et al. A TRNA Fragment, 5’-TiRNAVal, Suppresses the Wnt/β-Catenin Signaling Pathway by Targeting FZD3 in Breast Cancer. Cancer Lett. 2019, 457, 60–73. [Google Scholar] [CrossRef] [PubMed]
- Ormseth, M.J.; Wu, Q.; Zhao, S.; Allen, R.M.; Solus, J.; Sheng, Q.; Guo, Y.; Ye, F.; Ramirez-Solano, M.; Bridges, S.L.; et al. Circulating Microbial Small RNAs Are Altered in Patients with Rheumatoid Arthritis. Ann. Rheum. Dis. 2020, 79, 1557–1564. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Liu, L.; Han, S.; Wang, B.; Qin, J.; Bu, K.; Zhang, Y.; Li, Z.; Ma, L.; Tian, J.; et al. Expression of TiRNA and TRF in APP/PS1 Transgenic Mice and the Change of Related Proteins Expression. Ann. Transl. Med. 2021, 9, 1457. [Google Scholar] [CrossRef]
- Nätt, D.; Kugelberg, U.; Casas, E.; Nedstrand, E.; Zalavary, S.; Henriksson, P.; Nijm, C.; Jäderquist, J.; Sandborg, J.; Flinke, E.; et al. Human Sperm Displays Rapid Responses to Diet. PLoS Biol. 2019, 17, e3000559. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.; Shi, J.; Zhang, Y.; Zhang, H.; Liao, S.; Li, W.; Lei, L.; Han, C.; Ning, L.; Cao, Y.; et al. A Novel Class of TRNA-Derived Small RNAs Extremely Enriched in Mature Mouse Sperm. Cell Res. 2012, 22, 1609–1612. [Google Scholar] [CrossRef]
- Donkin, I.; Versteyhe, S.; Ingerslev, L.R.; Qian, K.; Mechta, M.; Nordkap, L.; Mortensen, B.; Appel, E.V.R.; Jørgensen, N.; Kristiansen, V.B.; et al. Obesity and Bariatric Surgery Drive Epigenetic Variation of Spermatozoa in Humans. Cell Metab. 2016, 23, 369–378. [Google Scholar] [CrossRef] [Green Version]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and Biological Role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017, 14, 1108–1123. [Google Scholar] [CrossRef] [Green Version]
- Hua, M.; Liu, W.; Chen, Y.; Zhang, F.; Xu, B.; Liu, S.; Chen, G.; Shi, H.; Wu, L. Identification of Small Non-Coding RNAs as Sperm Quality Biomarkers for in Vitro Fertilization. Cell Discov. 2019, 5, 20. [Google Scholar] [CrossRef] [Green Version]
- Keam, S.P.; Young, P.E.; McCorkindale, A.L.; Dang, T.H.Y.; Clancy, J.L.; Humphreys, D.T.; Preiss, T.; Hutvagner, G.; Martin, D.I.K.; Cropley, J.E.; et al. The Human Piwi Protein Hiwi2 Associates with TRNA-Derived PiRNAs in Somatic Cells. Nucleic Acids Res. 2014, 42, 8984–8995. [Google Scholar] [CrossRef]
- Kazimierczyk, M.; Jędroszkowiak, A.; Kowalczykiewicz, D.; Szymański, M.; Imiołczyk, B.; Ciesiołka, J.; Wrzesiński, J. TRNA-Derived Fragments from the Sus Scrofa Tissues Provide Evidence of Their Conserved Role in Mammalian Development. Biochem. Biophys. Res. Commun. 2019, 520, 514–519. [Google Scholar] [CrossRef]
- Tait, S.W.G.; Green, D.R. Mitochondria and Cell Death: Outer Membrane Permeabilization and Beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar]
- Saikia, M.; Jobava, R.; Parisien, M.; Putnam, A.; Krokowski, D.; Gao, X.-H.; Guan, B.-J.; Yuan, Y.; Jankowsky, E.; Feng, Z.; et al. Angiogenin-Cleaved TRNA Halves Interact with Cytochrome c, Protecting Cells from Apoptosis during Osmotic Stress. Mol. Cell. Biol. 2014, 34, 2450–2463. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Shen, J. Mucosal Immunity and TRNA, TRF, and TiRNA. J. Mol. Med. 2021, 99, 47–56. [Google Scholar] [CrossRef]
- Xie, Y.; Yao, L.; Yu, X.; Ruan, Y.; Li, Z.; Guo, J. Action Mechanisms and Research Methods of TRNA-Derived Small RNAs. Signal Transduct. Target. Ther. 2020, 5, 109. [Google Scholar] [CrossRef]
- Pandey, K.K.; Madhry, D.; Ravi Kumar, Y.S.; Malvankar, S.; Sapra, L.; Srivastava, R.K.; Bhattacharyya, S.; Verma, B. Regulatory Roles of TRNA-Derived RNA Fragments in Human Pathophysiology. Mol. Ther.-Nucleic Acids 2021, 26, 161–173. [Google Scholar]
- Pawar, K.; Shigematsu, M.; Sharbati, S.; Kirino, Y. Infection-Induced 50-Half Molecules of TRNAHisGUG Activate Toll-like Receptor 7. PLoS Biol. 2020, 18, e3000982. [Google Scholar] [CrossRef]
- Chiou, N.T.; Kageyama, R.; Ansel, K.M. Selective Export into Extracellular Vesicles and Function of TRNA Fragments during T Cell Activation. Cell Rep. 2018, 25, 3356–3370.e4. [Google Scholar] [CrossRef] [Green Version]
- Villarroya-Beltri, C.; Gutiérrez-Vázquez, C.; Sánchez-Cabo, F.; Pérez-Hernández, D.; Vázquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sánchez-Madrid, F. Sumoylated HnRNPA2B1 Controls the Sorting of MiRNAs into Exosomes through Binding to Specific Motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-Box Protein 1 Is Required to Sort MicroRNAs into Exosomes in Cells and in a Cell-Free Reaction. Elife 2016, 5, e19276. [Google Scholar] [CrossRef]
- McNab, F.W.; Ewbank, J.; Rajsbaum, R.; Stavropoulos, E.; Martirosyan, A.; Redford, P.S.; Wu, X.; Graham, C.M.; Saraiva, M.; Tsichlis, P.; et al. TPL-2–ERK1/2 Signaling Promotes Host Resistance against Intracellular Bacterial Infection by Negative Regulation of Type I IFN Production. J. Immunol. 2013, 191, 1732–1743. [Google Scholar] [CrossRef] [Green Version]
- vanLoosdregt, J.; Fleskens, V.; Tiemessen, M.M.; Mokry, M.; VanBoxtel, R.; Meerding, J.; Pals, C.E.G.M.; Kurek, D.; Baert, M.R.M.; Delemarre, E.M.; et al. Canonical Wnt Signaling Negatively Modulates Regulatory T Cell Function. Immunity 2013, 39, 298–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhahbi, J.M. 5′ TRNA Halves: The next Generation of Immune Signaling Molecules. Front. Immunol. 2015, 6, 74. [Google Scholar] [CrossRef] [Green Version]
- Samir, M.; Pessler, F. Small Non-Coding RNAs Associated with Viral Infectious Diseases of Veterinary Importance: Potential Clinical Applications. Front. Vet. Sci. 2016, 3, 22. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, Y.; Kong, L.; Song, C.; Chen, X.; Fang, X.; Zhang, C. TRNA-Derived RNA Fragments in the Exosomes of Bovine Milk and Colostrum. J. Food Compos. Anal. 2021, 102, 103948. [Google Scholar] [CrossRef]
- Liu, Q.Y.; Gu, X.R.; Zhao, Y.Q.; Zhang, J.; Zhao, Y.F.; Meng, Q.Y.; Xu, G.H.; Hu, X.X.; Li, N. Pig Large Tumor Suppressor 2 (Lats2), a Novel Gene That May Regulate the Fat Reduction in Adipocyte. BMB Rep. 2010, 43, 97–102. [Google Scholar] [CrossRef] [Green Version]
- Tao, E.-W.; Wang, H.-L.; Cheng, W.Y.; Liu, Q.-Q.; Chen, Y.-X.; Gao, Q.-Y. A Specific TRNA Half, 5′tiRNA-His-GTG, Responds to Hypoxia via the HIF1α/ANG Axis and Promotes Colorectal Cancer Progression by Regulating LATS2. J. Exp. Clin. Cancer Res. 2021, 40, 67. [Google Scholar] [CrossRef]
- Bordoni, L.; Gabbianelli, R. The Neglected Nutrigenomics of Milk: What Is the Role of Inter-Species Transfer of Small Non-Coding RNA? Food Biosci. 2021, 39, 100796. [Google Scholar] [CrossRef]
- Taxis, T.M.; Bauermann, F.V.; Ridpath, J.F.; Casas, E. Analysis of TRNA Halves (TsRNAs) in Serum from Cattle Challenged with Bovine Viral Diarrhea Virus. Genet. Mol. Biol. 2019, 42, 374–379. [Google Scholar] [CrossRef]
- Ivanov, P. Emerging Roles of TRNA-Derived Fragments in Viral Infections: The Case of Respiratory Syncytial Virus. Mol. Ther. 2015, 23, 1557–1558. [Google Scholar] [CrossRef]
- Taxis, T.M.; Kehrli, M.E.; D’Orey-Branco, R.; Casas, E. Association of Transfer RNA Fragments in White Blood Cells with Antibody Response to Bovine Leukemia Virus in Holstein Cattle. Front. Genet. 2018, 9, 236. [Google Scholar] [CrossRef]
- Casas, E.; Cai, G.; Kuehn, L.A.; Register, K.B.; McDaneld, T.G.; Neill, J.D. Association of Circulating Transfer RNA Fragments with Antibody Response to Mycoplasma Bovis in Beef Cattle. BMC Vet. Res. 2018, 14, 89. [Google Scholar] [CrossRef]
- Fricker, R.; Brogli, R.; Luidalepp, H.; Wyss, L.; Fasnacht, M.; Joss, O.; Zywicki, M.; Helm, M.; Schneider, A.; Cristodero, M.; et al. A TRNA Half Modulates Translation as Stress Response in Trypanosoma Brucei. Nat. Commun. 2019, 10, 118. [Google Scholar] [CrossRef] [Green Version]
- Hotamisligil, G.S.; Davis, R.J. Cell Signaling and Stress Responses. Cold Spring Harb. Perspect. Biol. 2016, 8, a006072. [Google Scholar] [CrossRef] [Green Version]
- Campos-Melo, D.; Hawley, Z.C.E.; Droppelmann, C.A.; Strong, M.J. The Integral Role of RNA in Stress Granule Formation and Function. Front. Cell Dev. Biol. 2021, 9, 808. [Google Scholar] [CrossRef]
- Haack, F.; Trakooljul, N.; Gley, K.; Murani, E.; Hadlich, F.; Wimmers, K.; Ponsuksili, S. Deep Sequencing of Small Non-Coding RNA Highlights Brain-Specific Expression Patterns and RNA Cleavage. RNA Biol. 2019, 16, 1764–1774. [Google Scholar] [CrossRef]
- Gley, K.; Hadlich, F.; Trakooljul, N.; Haack, F.; Murani, E.; Gimsa, U.; Wimmers, K.; Ponsuksili, S. Multi-Transcript Level Profiling Revealed Distinct MRNA, MiRNA, and TRNA-Derived Fragment Bio-Signatures for Coping Behavior Linked Haplotypes in HPA Axis and Limbic System. Front. Genet. 2021, 12, 1558. [Google Scholar] [CrossRef]
- Jehn, J.; Treml, J.; Wulsch, S.; Ottum, B.; Erb, V.; Hewel, C.; Kooijmans, R.N.; Wester, L.; Fast, I.; Rosenkranz, D. 5′ TRNA Halves Are Highly Expressed in the Primate Hippocampus and Might Sequence-Specifically Regulate Gene Expression. RNA 2020, 26, 694–707. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Kuscu, C.; Dutta, A. Biogenesis and Function of Transfer RNA-Related Fragments (TRFs). Trends Biochem. Sci. 2016, 41, 679–689. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Mudunuri, S.B.; Anaya, J.; Dutta, A. TRFdb: A Database for Transfer RNA Fragments. Nucleic Acids Res. 2015, 43, D141–D145. [Google Scholar] [CrossRef]
- la Ferlita, A.; Alaimo, S.; Veneziano, D.; Nigita, G.; Balatti, V.; Croce, C.M.; Ferro, A.; Pulvirenti, A. Identification of TRNA-Derived NcRNAs in TCGA and NCI-60 Panel Cell Lines and Development of the Public Database TRFexplorer. Database 2019, 2019, 115. [Google Scholar] [CrossRef]
- Zuo, Y.; Zhu, L.; Guo, Z.; Liu, W.; Zhang, J.; Zeng, Z.; Wu, Q.; Cheng, J.; Fu, X.; Jin, Y.; et al. TsRBase: A Comprehensive Database for Expression and Function of TsRNAs in Multiple Species. Nucleic Acids Res. 2021, 49, D1038–D1045. [Google Scholar] [CrossRef]
- Chan, P.P.; Lowe, T.M. GtRNAdb 2.0: An Expanded Database of Transfer RNA Genes Identified in Complete and Draft Genomes. Nucleic Acids Res. 2016, 44, D184–D189. [Google Scholar] [CrossRef] [Green Version]
- Dhahbi, J.M. Circulating Small Noncoding RNAs as Biomarkers of Aging. Ageing Res. Rev. 2014, 17, 86–98. [Google Scholar] [CrossRef]
- Miretti, S.; Lecchi, C.; Ceciliani, F.; Baratta, M. MicroRNAs as Biomarkers for Animal Health and Welfare in Livestock. Front. Vet. Sci. 2020, 7, 985. [Google Scholar] [CrossRef]
- Fleming, D.S.; Miller, L.C. Differentially Expressed MiRNAs and TRNA Genes Affect Host Homeostasis during Highly Pathogenic Porcine Reproductive and Respiratory Syndrome Virus Infections in Young Pigs. Front. Genet. 2019, 10, 691. [Google Scholar] [CrossRef] [Green Version]







| Class | Symbol | Characteristics | Biological Functions Associations | Ref. | |
|---|---|---|---|---|---|
| Small non-coding RNAs (sncRNAs) | MicroRNAs | miRNAs | 18–26 nt; comprises 2% of human genome and regulate up to 50% of protein-coding genes. | Control of proliferation, apoptosis, and differentiation. | [9] |
| Small interfering RNAs | siRNA | 19–23 nt; processed by Dicer, and guide sequence-specific degradation of target mRNA. | Post-transcriptional regulation of gene expression. | [10] | |
| Piwi-interacting RNAs | piRNAs | 24–31 nt; made by single-stranded RNA (ssRNA) precursors and it is Dicer-independent. | Embryonic development, germline DNA integrity, transposon transcription silencing, translation suppression, heterochromatin creation, and sex determination epigenetic control. | [11] | |
| Small nucleolar RNAs | snoRNAs | 60–300 nt; divided into two classes: C/D box snoRNAs and H/ACA box snoRNAs. It is primarily accumulated in the nucleoli. | Responsible for post-transcriptional modification and maturation of ribosomal RNAs (rRNAs), small nuclear RNAs (snRNAs), and other RNAs (snRNAs). | [12] | |
| Centromere repeat-associated small interacting RNAs | crasiRNAs | 34–42 nt; processed from long dsRNAs. | Activation of heterochromatin and centromeric proteins. | [13] | |
| Telomere-specific small RNAs | tel-sRNAs | ~24 nt; pi-like small RNA and independent of Dicer processing. | Epigenetic regulation. | [14] | |
| Pyknons | >16 nt long; observed in groups in intergenic and intronic domains. | Primarily engaged in cell communication, transcriptional regulation, signalling, and transport. | [15] | ||
| tRNA fragments | tRFs | 14–30 nt; dependent on angiogenin and Dicer processing. | Diverse molecular and physiological processes, including gene suppression, RNA processing, protein translation, stress responses, cell proliferation, and differentiation. | [16] | |
| tRNA-derived stress-induced RNAs | tiRNAs (tRNA halves) | 29–50 nt; the most abundant right downstream of transcriptional end sites. It exhibits spatial preservation patterns and predominantly resides in GC-rich promoters. | Control of protein-coding gene transcription by targeting epigenetic silencing complexes. | [17] |
| Species | ANG | Protein Length | Ref. |
|---|---|---|---|
| Human (H. sapiens) | 1 | 147 aa | [49] |
| Mouse (M. musculus) | 5 (3 pseudogenes) | 145 aa | [49] |
| Rat (R. norvegicus) | 2 | 145 aa | [49] |
| Dog (C. lupus familiaris) | 1 | 145 aa | [50] |
| Cattle (B. Taurus) | 3 | 148 aa | [50] |
| Pig (S. scrofa) | 2 | 202 aa | [51] |
| Rabbit (O. cuniculus) | 1 | 149 aa | [51] |
| Rainbow trout (O. mykiss) | 6 | 205 aa | [52] |
| Zebrafish (D. rerio) | 5 | 149 aa | [52] |
|
Aging Hallmarks | tiRNA ID | tiRNA Type | Gene or Protein Target | Mechanism | Ref. |
|---|---|---|---|---|---|
| Cellular senescence | tiRNA-5034-GluTTC-2 | 5′-tiRNA | - | Downregulated in cancer tissue, and the degree of expression is inversely related to tumour growth. | [79] |
| tiRNALys | 5′-tiRNA | - | Regulation of aging hallmarks | [78] | |
| Loss of proteostasis | tiRNAAla | 5′-tiRNA | elF2a | Inhibits protein synthesis | [80] |
| tiRNACys | 5′-tiRNA | elF2a | Inhibits protein synthesis | [80] | |
| tiRNAVal | 5′-tiRNA | FZD3 | Regulation of WNT/β-Catenin pathway | [81] | |
| Aging related diseases | tiRNAAsnGTT tiRNAIleAAT tiRNAAspGTC | 5′-tiRNA | - | Downregulation observed in rheumatoid arthritis patients | [82] |
| tRNAVal derived | 5′-tiRNA 3′-tiRNA | Aβ/Tau | May inhibit Aβ production and Tau protein hyperphosphorylation in Alzheimer’s disease | [83] |
| Disease | tiRNA ID | tiRNA Type | Mechanism | Ref. |
|---|---|---|---|---|
| Bovine viral diarrhoea virus (BVDV) | tiRNaGlyCCC tiRNAGlyGCC | 5′-tiRNA | It is downregulated, it may define the immune response against BVDV | [108] |
| Respiratory syncytial virus (RSV) | tiRNAGluCTC tiRNAGlu | 5′-tiRNA | RSV promotes ANG expression, and then cleaves specific tRNAs that may inhibit APOER2 production | [109] |
| Bovine Leukemia Virus (BLV) | tiRNAGlnCTG tiRNAGlnTTG tiRNAHisGTG | 5′-tiRNA | It targets white blood cells causing dysregulated immune functions and immunosuppression. | [110] |
| Mycoplasma bovis | tiRNASelCysUGA | 5′-tiRNA | It may be correlated with a host defence mechanism enhanced by bacterial infection. | [111] |
| Trypanosoma brucei (rHAT) | tiRNAThr | 3′-tiRNA | During the stress recovery phase, it attaches to the ribosome and increases protein production. | [112] |
| Organism | tRNA Decoding Standard 20AA | TCA Suppressor tRNAs | Genome Version |
|---|---|---|---|
| H. sapiens | 415 | 1 | Feb.2009 GRCh37/hg19 |
| S. scrofa | 471 | 1 | Feb.2017 Sscrofa11.1 |
| G. gallus | 280 | 1 | Mar.2018 GRCg6a |
| B. taurus | 619 | 1 | Apr.2018 ARS-UCD1.2/bosTau9 |
| Name | Description | Link (accessed on 9 April 2022) |
|---|---|---|
| tRFexplorer | Publicly accessible database that allows users to view the expression profiles of tRNA-derived ncRNAs in each NCI-60 cell line. | https://trfexplorer.cloud/ |
| tRFdb | The first database of transfer RNA fragments (tRFs) | http://genome.bioch.virginia.edu/trfdb/ |
| tsRBase | Multi-species database of tsRNA sequences, expression characteristics, and function. | http://www.tsrbase.org/ |
| GtRNAdb v2 | tRNA gene predictions on complete or nearly complete genomes. | http://gtrnadb.ucsc.edu/GtRNAdb2/ |
| tDRnamer | Standardised naming for tRNA-derived RNAs | http://trna.ucsc.edu/tDRnamer/ |
| tRAX | In-depth analysis of tRNA-derived small RNAs (tDRs), mature tRNAs, and RNA modification inference from high-throughput small RNA sequencing data. | http://trna.ucsc.edu/tRAX |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarais, F.; Perdomo-Sabogal, A.; Wimmers, K.; Ponsuksili, S. tiRNAs: Insights into Their Biogenesis, Functions, and Future Applications in Livestock Research. Non-Coding RNA 2022, 8, 37. https://doi.org/10.3390/ncrna8030037
Sarais F, Perdomo-Sabogal A, Wimmers K, Ponsuksili S. tiRNAs: Insights into Their Biogenesis, Functions, and Future Applications in Livestock Research. Non-Coding RNA. 2022; 8(3):37. https://doi.org/10.3390/ncrna8030037
Chicago/Turabian StyleSarais, Fabio, Alvaro Perdomo-Sabogal, Klaus Wimmers, and Siriluck Ponsuksili. 2022. "tiRNAs: Insights into Their Biogenesis, Functions, and Future Applications in Livestock Research" Non-Coding RNA 8, no. 3: 37. https://doi.org/10.3390/ncrna8030037
APA StyleSarais, F., Perdomo-Sabogal, A., Wimmers, K., & Ponsuksili, S. (2022). tiRNAs: Insights into Their Biogenesis, Functions, and Future Applications in Livestock Research. Non-Coding RNA, 8(3), 37. https://doi.org/10.3390/ncrna8030037