RNA-Mediated Regulation of Meiosis in Budding Yeast
Abstract
:1. Introduction
2. Non-Coding RNA
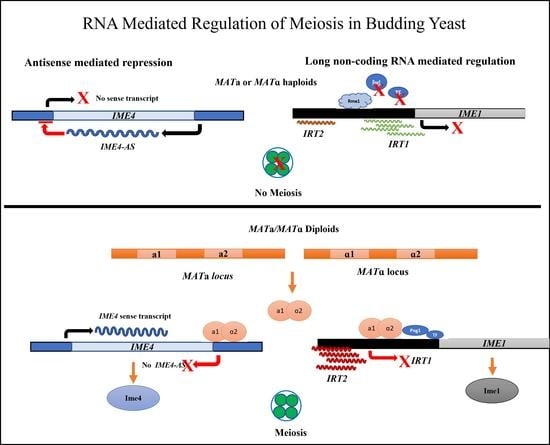
3. Meiotic Non-Coding RNA
4. RNA Processing Factors Regulate Meiosis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Haber, J.E.; Halvorson, H.O. Chapter 2 Regulation of Sporulation in Yeast. Curr. Top. Dev. Biol. 1972, 7, 61–83. [Google Scholar]
- Roth, R.; Fogel, S. A system selective for yeast mutants deficient in meiotic recombination. Mol. Gen. Genet. MGG 1971, 112, 295–305. [Google Scholar] [CrossRef]
- Yamamoto, M. The selective elimination of messenger RNA underlies the mitosis-meiosis switch in fission yeast. Proc. Jpn. Acad. Ser. B 2010, 86, 788–797. [Google Scholar] [CrossRef] [Green Version]
- Kassir, Y.; Simchen, G. Regulation of mating and meiosis in yeast by the mating-type region. Genetics 1976, 82, 187–206. [Google Scholar] [CrossRef]
- Rine, J.; Sprague, G.F.; Herskowitz, I. rme1 Mutation of Saccharomyces cerevisiae: Map position and bypass of mating type locus control of sporulation. Mol. Cell. Biol. 1981, 1, 958–960. [Google Scholar]
- Mitchell, A.P.; Herskowitz, I. Activation of meiosis and sporulation by repression of the RME1 product in yeast. Nature 1986, 319, 738–742. [Google Scholar] [CrossRef]
- Smith, H.E.; Mitchell, A.P. A transcriptional cascade governs entry into meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1989, 9, 2142–2152. [Google Scholar]
- Kassir, Y.; Granot, D.; Simchen, G. IME1, a positive regulator gene of meiosis in S. cerevisiae. Cell 1988, 52, 853–862. [Google Scholar]
- Toone, W.M.; Johnson, A.L.; Banks, G.R.; Toyn, J.H.; Stuart, D.; Wittenberg, C.; Johnston, L.H. Rme1, a negative regulator of meiosis, is also a positive activator of G1 cyclin gene expression. EMBO J. 1995, 14, 5824–5832. [Google Scholar] [CrossRef]
- Covitz, P.A.; Mitchell, A.P. Repression by the yeast meiotic inhibitor RME1. Genes Dev. 1993, 7, 1598–1608. [Google Scholar]
- Bushkin, G.G.; Pincus, D.; Morgan, J.T.; Richardson, K.; Lewis, C.; Chan, S.H.; Bartel, D.P.; Fink, G.R. m6A modification of a 3′ UTR site reduces RME1 mRNA levels to promote meiosis. Nat. Commun. 2019, 10, 3414. [Google Scholar] [CrossRef] [Green Version]
- Shah, J.C.; Clancy, M.J. IME4, a gene that mediates MAT and nutritional control of meiosis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1992, 12, 1078–1086. [Google Scholar]
- Tisseur, M.; Kwapisz, M.; Morillon, A. Pervasive transcription—Lessons from yeast. Biochimie 2011, 93, 1889–1896. [Google Scholar] [CrossRef]
- Tudek, A.; Candelli, T.; Libri, D. Non-coding transcription by RNA polymerase II in yeast: Hasard or nécessité? Biochimie 2015, 117, 28–36. [Google Scholar]
- Zhang, P.; Wu, W.; Chen, Q.; Chen, M. Non-Coding RNAs and their Integrated Networks. J. Integr. Bioinform. 2019, 1, 20190027. [Google Scholar]
- Sana, J.; Faltejskova, P.; Svoboda, M.; Slaby, O. Novel classes of non-coding RNAs and cancer. J. Transl. Med. 2012, 10, 103. [Google Scholar]
- Xu, Z.; Wei, W.; Gagneur, J.; Perocchi, F.; Clauder-Münster, S.; Camblong, J.; Guffanti, E.; Stutz, F.; Huber, W.; Steinmetz, L.M. Bidirectional promoters generate pervasive transcription in yeast. Nature 2009, 457, 1033–1037. [Google Scholar] [CrossRef] [Green Version]
- Preker, P.; Nielsen, J.; Schierup, M.H.; Heick Jensen, T. RNA polymerase plays both sides: Vivid and bidirectional transcription around and upstream of active promoters. Cell Cycle 2009, 8, 1105–1111. [Google Scholar] [CrossRef] [Green Version]
- Nevers, A.; Doyen, A.; Malabat, C.; Néron, B.; Kergrohen, T.; Jacquier, A.; Badis, G. Antisense transcriptional interference mediates condition-specific gene repression in budding yeast. Nucleic Acids Res. 2018, 46, 6009–6025. [Google Scholar] [CrossRef]
- Neil, H.; Malabat, C.; D’Aubenton-Carafa, Y.; Xu, Z.; Steinmetz, L.M.; Jacquier, A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 2009, 457, 1038–1042. [Google Scholar]
- Marquardt, S.; Hazelbaker, D.Z.; Buratowski, S. Distinct RNA degradation pathways and 3’ extensions of yeast non-coding RNA species. Transcription 2011, 2, 145–154. [Google Scholar] [CrossRef] [Green Version]
- van Dijk, E.L.; Chen, C.L.; D’Aubenton-Carafa, Y.; Gourvennec, S.; Kwapisz, M.; Roche, V.; Bertrand, C.; Silvain, M.; Legoix-Né, P.; Loeillet, S.; et al. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 2011, 475, 114–117. [Google Scholar] [CrossRef]
- Schulz, D.; Schwalb, B.; Kiesel, A.; Baejen, C.; Torkler, P.; Gagneur, J.; Soeding, J.; Cramer, P. Transcriptome Surveillance by Selective Termination of Noncoding RNA Synthesis. Cell 2013, 155, 1075–1087. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 925–933. [Google Scholar] [CrossRef] [Green Version]
- Hombach, S.; Kretz, M. Non-coding RNAs: Classification, Biology and Functioning. In Non-Coding RNAs in Colorectal Cancer; Springer: Cham, Swizherland, 2016; pp. 3–17. [Google Scholar]
- Ernst, C.; Morton, C.C. Identification and function of long non-coding RNA. Front. Cell. Neurosci. 2013, 7, 168. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, W.; Zheng, K.; Liu, Z.-F. Small Non-Coding RNAs: New Insights in Modulation of Host Immune Response by Intracellular Bacterial Pathogens. Front. Immunol. 2016, 7, 431. [Google Scholar]
- Guillier, M.; Gottesman, S. The 5′ end of two redundant sRNAs is involved in the regulation of multiple targets, including their own regulator. Nucleic Acids Res. 2008, 36, 6781–6794. [Google Scholar] [CrossRef] [Green Version]
- Ordas, A.; Kanwal, Z.; Lindenberg, V.; Rougeot, J.; Mink, M.; Spaink, H.P.; Meijer, A.H. MicroRNA-146 function in the innate immune transcriptome response of zebrafish embryos to Salmonella typhimurium infection. BMC Genom. 2013, 14, 696. [Google Scholar] [CrossRef] [Green Version]
- Tycowski, K.T.; Guo, Y.E.; Lee, N.; Moss, W.N.; Vallery, T.K.; Xie, M.; Steitz, J.A. Viral noncoding RNAs: More surprises. Genes Dev. 2015, 29, 567–584. [Google Scholar] [CrossRef] [Green Version]
- Clemson, C.M.; McNeil, J.A.; Willard, H.F.; Lawrence, J.B. XIST RNA paints the inactive X chromosome at interphase: Evidence for a novel RNA involved in nuclear/chromosome structure. J. Cell Biol. 1996, 132, 259–275. [Google Scholar] [CrossRef] [Green Version]
- Szachnowski, U.; Andjus, S.; Foretek, D.; Morillon, A.; Wery, M. Endogenous RNAi pathway evolutionarily shapes the destiny of the antisense lncRNAs transcriptome. Life Sci. Alliance 2019, 2, e201900407. [Google Scholar] [CrossRef]
- Atkinson, S.R.; Marguerat, S.; Bitton, D.A.; Rodríguez-López, M.; Rallis, C.; Lemay, J.-F.; Cotobal, C.; Malecki, M.; Smialowski, P.; Mata, J.; et al. Long noncoding RNA repertoire and targeting by nuclear exosome, cytoplasmic exonuclease, and RNAi in fission yeast. RNA 2018, 24, 1195–1213. [Google Scholar] [CrossRef] [Green Version]
- Han, P.; Chang, C.-P. Long non-coding RNA and chromatin remodeling. RNA Biol. 2015, 12, 1094–1098. [Google Scholar] [CrossRef] [Green Version]
- Dinger, M.E.; Amaral, P.P.; Mercer, T.R.; Pang, K.C.; Bruce, S.J.; Gardiner, B.B.; Askarian-Amiri, M.E.; Ru, K.; Soldà, G.; Simons, C.; et al. Long noncoding RNAs in mouse embryonic stem cell pluripotency and differentiation. Genome Res. 2008, 18, 1433–1445. [Google Scholar] [CrossRef] [Green Version]
- Sheik Mohamed, J.; Gaughwin, P.M.; Lim, B.; Robson, P.; Lipovich, L. Conserved long noncoding RNAs transcriptionally regulated by Oct4 and Nanog modulate pluripotency in mouse embryonic stem cells. RNA 2010, 16, 324–337. [Google Scholar] [CrossRef] [Green Version]
- Fatica, A.; Bozzoni, I. Long non-coding RNAs: New players in cell differentiation and development. Nat. Rev. Genet. 2014, 15, 7–21. [Google Scholar] [CrossRef]
- Yassour, M.; Pfiffner, J.; Levin, J.Z.; Adiconis, X.; Gnirke, A.; Nusbaum, C.; Thompson, D.-A.; Friedman, N.; Regev, A. Strand-specific RNA sequencing reveals extensive regulated long antisense transcripts that are conserved across yeast species. Genome Biol. 2010, 11, R87. [Google Scholar] [CrossRef] [Green Version]
- Rhind, N.; Chen, Z.; Yassour, M.; Thompson, D.A.; Haas, B.J.; Habib, N.; Wapinski, I.; Roy, S.; Lin, M.F.; Heiman, D.I.; et al. Comparative Functional Genomics of the Fission Yeasts. Science 2011, 332, 930–936. [Google Scholar] [CrossRef] [Green Version]
- Churchman, L.S.; Weissman, J.S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 2011, 469, 368–373. [Google Scholar] [CrossRef] [Green Version]
- Eser, P.; Wachutka, L.; Maier, K.C.; Demel, C.; Boroni, M.; Iyer, S.; Cramer, P.; Gagneur, J. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. Mol. Syst. Biol. 2016, 12, 857. [Google Scholar] [CrossRef]
- Bitton, D.A.; Grallert, A.; Scutt, P.J.; Yates, T.; Li, Y.; Bradford, J.R.; Hey, Y.; Pepper, S.D.; Hagan, I.M.; Miller, C.J. Programmed fluctuations in sense/antisense transcript ratios drive sexual differentiation in S. pombe. Mol. Syst. Biol. 2011, 7, 559. [Google Scholar] [CrossRef] [PubMed]
- Wery, M.; Gautier, C.; Descrimes, M.; Yoda, M.; Vennin-Rendos, H.; Migeot, V.; Gautheret, D.; Hermand, D.; Morillon, A. Native elongating transcript sequencing reveals global anti-correlation between sense and antisense nascent transcription in fission yeast. RNA 2018, 24, 196–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lardenois, A.; Liu, Y.; Walther, T.; Chalmel, F.; Evrard, B.; Granovskaia, M.; Chu, A.; Davis, R.W.; Steinmetz, L.M.; Primig, M. Execution of the meiotic noncoding RNA expression program and the onset of gametogenesis in yeast require the conserved exosome subunit Rrp6. Proc. Natl. Acad. Sci. USA 2011, 108, 1058–1063. [Google Scholar] [CrossRef] [Green Version]
- Harigaya, Y.; Tanaka, H.; Yamanaka, S.; Tanaka, K.; Watanabe, Y.; Tsutsumi, C.; Chikashige, Y.; Hiraoka, Y.; Yamashita, A.; Yamamoto, M. Selective elimination of messenger RNA prevents an incidence of untimely meiosis. Nature 2006, 442, 45–50. [Google Scholar] [CrossRef]
- St-André, O.; Lemieux, C.; Perreault, A.; Lackner, D.H.; Bähler, J.; Bachand, F. Negative Regulation of Meiotic Gene Expression by the Nuclear Poly(a)-binding Protein in Fission Yeast. J. Biol. Chem. 2010, 285, 27859–27868. [Google Scholar] [CrossRef] [Green Version]
- David, L.; Huber, W.; Granovskaia, M.; Toedling, J.; Palm, C.J.; Bofkin, L.; Jones, T.; Davis, R.W.; Steinmetz, L.M. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA 2006, 103, 5320–5325. [Google Scholar] [CrossRef] [Green Version]
- Kim Guisbert, K.S.; Zhang, Y.; Flatow, J.; Hurtado, S.; Staley, J.P.; Lin, S.; Sontheimer, E.J. Meiosis-induced alterations in transcript architecture and noncoding RNA expression in S. cerevisiae. RNA 2012, 18, 1142–1153. [Google Scholar] [CrossRef] [Green Version]
- Becker, E.; Com, E.; Lavigne, R.; Guilleux, M.-H.; Evrard, B.; Pineau, C.; Primig, M. The protein expression landscape of mitosis and meiosis in diploid budding yeast. J. Proteomics 2017, 156, 5–19. [Google Scholar] [CrossRef]
- Hongay, C.F.; Grisafi, P.L.; Galitski, T.; Fink, G.R. Antisense transcription controls cell fate in Saccharomyces cerevisiae. Cell 2006, 127, 735–745. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Ma, H.; Pugh, B.F. Stable and dynamic nucleosome states during a meiotic developmental process. Genome Res. 2011, 21, 875–884. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-M.; Neiman, A.M. A conserved regulatory role for antisense RNA in meiotic gene expression in yeast. Curr. Opin. Microbiol. 2011, 14, 655–659. [Google Scholar] [CrossRef] [PubMed]
- van Werven, F.J.; Neuert, G.; Hendrick, N.; Lardenois, A.; Buratowski, S.; van Oudenaarden, A.; Primig, M.; Amon, A. Transcription of Two Long Noncoding RNAs Mediates Mating-Type Control of Gametogenesis in Budding Yeast. Cell 2012, 150, 1170–1181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, T.; Buratowski, S. Dimethylation of H3K4 by Set1 Recruits the Set3 Histone Deacetylase Complex to 5′ Transcribed Regions. Cell 2009, 137, 259–272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keogh, M.-C.; Kurdistani, S.K.; Morris, S.A.; Ahn, S.H.; Podolny, V.; Collins, S.R.; Schuldiner, M.; Chin, K.; Punna, T.; Thompson, N.J.; et al. Cotranscriptional Set2 Methylation of Histone H3 Lysine 36 Recruits a Repressive Rpd3 Complex. Cell 2005, 123, 593–605. [Google Scholar] [CrossRef] [Green Version]
- Deutschbauer, A.M.; Davis, R.W. Quantitative trait loci mapped to single-nucleotide resolution in yeast. Nat. Genet. 2005, 37, 1333–1340. [Google Scholar] [CrossRef]
- Gerke, J.; Lorenz, K.; Cohen, B. Genetic Interactions Between Transcription Factors Cause Natural Variation in Yeast. Science 2009, 323, 498–501. [Google Scholar] [CrossRef] [Green Version]
- Moretto, F.; Wood, N.E.; Chia, M.; Li, C.; Luscombe, N.M.; van Werven, F.J. Transcription levels of a noncoding RNA orchestrate opposing regulatory and cell fate outcomes in yeast. Cell Rep. 2021, 34, 108643. [Google Scholar] [CrossRef]
- Moretto, F.; Wood, N.E.; Kelly, G.; Doncic, A.; van Werven, F.J. A regulatory circuit of two lncRNAs and a master regulator directs cell fate in yeast. Nat. Commun. 2018, 9, 780. [Google Scholar] [CrossRef] [Green Version]
- Bernier, M.; Luo, Y.; Nwokelo, K.C.; Goodwin, M.; Dreher, S.J.; Zhang, P.; Parthun, M.R.; Fondufe-Mittendorf, Y.; Ottesen, J.J.; Poirier, M.G. Linker histone H1 and H3K56 acetylation are antagonistic regulators of nucleosome dynamics. Nat. Commun. 2015, 6, 10152. [Google Scholar] [CrossRef] [Green Version]
- Parker, S.; Fraczek, M.G.; Wu, J.; Shamsah, S.; Manousaki, A.; Dungrattanalert, K.; de Almeida, R.A.; Invernizzi, E.; Burgis, T.; Omara, W.; et al. Large-scale profiling of noncoding RNA function in yeast. PLoS Genet. 2018, 14, e1007253. [Google Scholar] [CrossRef] [Green Version]
- Andric, V.; Nevers, A.; Hazra, D.; Auxilien, S.; Menant, A.; Graille, M.; Palancade, B.; Rougemaille, M. A scaffold lncRNA shapes the mitosis to meiosis switch. Nat. Commun. 2021, 12, 770. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, K.; Futcher, B.; Leatherwood, J. mmi1 and rep2 mRNAs are novel RNA targets of the Mei2 RNA-binding protein during early meiosis in Schizosaccharomyces pombe. Open Biol. 2018, 8, 180110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugiyama, T.; Sugioka-Sugiyama, R.; Hada, K.; Niwa, R. Rhn1, a nuclear protein, is required for suppression of meiotic mRNAs in mitotically dividing fission yeast. PLoS ONE 2012, 7, e42962. [Google Scholar] [CrossRef] [PubMed]
- Andric, V.; Rougemaille, M. Long Non-Coding RNAs in the Control of Gametogenesis: Lessons from Fission Yeast. Non-Coding RNA 2021, 7, 34. [Google Scholar] [CrossRef]
- Kilchert, C.; Wittmann, S.; Passoni, M.; Shah, S.; Granneman, S.; Vasiljeva, L. Regulation of mRNA Levels by Decay-Promoting Introns that Recruit the Exosome Specificity Factor Mmi1. Cell Rep. 2015, 13, 2504–2515. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.-M.; Futcher, B.; Leatherwood, J. The Fission Yeast RNA Binding Protein Mmi1 Regulates Meiotic Genes by Controlling Intron Specific Splicing and Polyadenylation Coupled RNA Turnover. PLoS ONE 2011, 6, e26804. [Google Scholar] [CrossRef] [Green Version]
- Hiriart, E.; Vavasseur, A.; Touat-Todeschini, L.; Yamashita, A.; Gilquin, B.; Lambert, E.; Perot, J.; Shichino, Y.; Nazaret, N.; Boyault, C.; et al. Mmi1 RNA surveillance machinery directs RNAi complex RITS to specific meiotic genes in fission yeast. EMBO J. 2012, 31, 2296–2308. [Google Scholar] [CrossRef]
- Shichino, Y.; Yamashita, A.; Yamamoto, M. Meiotic long non-coding meiRNA accumulates as a dot at its genetic locus facilitated by Mmi1 and plays as a decoy to lure Mmi1. Open Biol. 2014, 4, 140022. [Google Scholar] [CrossRef] [Green Version]
- Touat-Todeschini, L.; Shichino, Y.; Dangin, M.; Thierry-Mieg, N.; Gilquin, B.; Hiriart, E.; Sachidanandam, R.; Lambert, E.; Brettschneider, J.; Reuter, M.; et al. Selective termination of lnc RNA transcription promotes heterochromatin silencing and cell differentiation. EMBO J. 2017, 36, 2626–2641. [Google Scholar] [CrossRef]
- Zofall, M.; Yamanaka, S.; Reyes-Turcu, F.E.; Zhang, K.; Rubin, C.; Grewal, S.I.S. RNA Elimination Machinery Targeting Meiotic mRNAs Promotes Facultative Heterochromatin Formation. Science 2012, 335, 96–100. [Google Scholar] [CrossRef]
- Holm, L.R.; Thon, G. New romance between RNA degradation pathways: Mmi1 and RNAi meet on heterochromatic islands. EMBO J. 2012, 31, 2242–2243. [Google Scholar] [CrossRef]
- Lee, S.Y.; Hung, S.; Esnault, C.; Pathak, R.; Johnson, K.R.; Bankole, O.; Yamashita, A.; Zhang, H.; Levin, H.L. Dense Transposon Integration Reveals Essential Cleavage and Polyadenylation Factors Promote Heterochromatin Formation. Cell Rep. 2020, 30, 2686–2698.e8. [Google Scholar] [CrossRef] [Green Version]
- Chalamcharla, V.R.; Folco, H.D.; Dhakshnamoorthy, J.; Grewal, S.I.S. Conserved factor Dhp1/Rat1/Xrn2 triggers premature transcription termination and nucleates heterochromatin to promote gene silencing. Proc. Natl. Acad. Sci. USA 2015, 112, 15548–15555. [Google Scholar] [CrossRef] [Green Version]
- Srividya, I.; Tirupataiah, S.; Mishra, K. Yeast Transcription Termination Factor Rtt103 Functions in DNA Damage Response. PLoS ONE 2012, 7, e31288. [Google Scholar] [CrossRef] [Green Version]
- Kloimwieder, A.; Winston, F. A Screen for Germination Mutants in Saccharomyces cerevisiae. G3 2011, 1, 143–149. [Google Scholar] [CrossRef] [Green Version]
- Nemec, C.M.; Yang, F.; Gilmore, J.M.; Hintermair, C.; Ho, Y.-H.; Tseng, S.C.; Heidemann, M.; Zhang, Y.; Florens, L.; Gasch, A.P.; et al. Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner. Proc. Natl. Acad. Sci. USA 2017, 114, E3944–E3953. [Google Scholar] [CrossRef] [Green Version]
- Ding, D.-Q.; Okamasa, K.; Katou, Y.; Oya, E.; Nakayama, J.; Chikashige, Y.; Shirahige, K.; Haraguchi, T.; Hiraoka, Y. Chromosome-associated RNA–protein complexes promote pairing of homologous chromosomes during meiosis in Schizosaccharomyces pombe. Nat. Commun. 2019, 10, 5598. [Google Scholar] [CrossRef] [Green Version]
- Wery, M.; Descrimes, M.; Vogt, N.; Dallongeville, A.-S.; Gautheret, D.; Morillon, A. Nonsense-Mediated Decay Restricts LncRNA Levels in Yeast Unless Blocked by Double-Stranded RNA Structure. Mol. Cell 2016, 61, 379–392. [Google Scholar] [CrossRef] [Green Version]
- Malabat, C.; Feuerbach, F.; Ma, L.; Saveanu, C.; Jacquier, A. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. Elife 2015, 4, e06722. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pondugala, V.V.; Mishra, K. RNA-Mediated Regulation of Meiosis in Budding Yeast. Non-Coding RNA 2022, 8, 77. https://doi.org/10.3390/ncrna8060077
Pondugala VV, Mishra K. RNA-Mediated Regulation of Meiosis in Budding Yeast. Non-Coding RNA. 2022; 8(6):77. https://doi.org/10.3390/ncrna8060077
Chicago/Turabian StylePondugala, Vidya Vardhini, and Krishnaveni Mishra. 2022. "RNA-Mediated Regulation of Meiosis in Budding Yeast" Non-Coding RNA 8, no. 6: 77. https://doi.org/10.3390/ncrna8060077
APA StylePondugala, V. V., & Mishra, K. (2022). RNA-Mediated Regulation of Meiosis in Budding Yeast. Non-Coding RNA, 8(6), 77. https://doi.org/10.3390/ncrna8060077