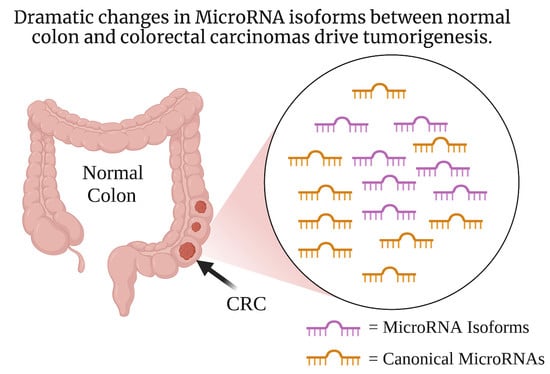
A Review of IsomiRs in Colorectal Cancer
Abstract
:1. Introduction
1.1. Initial Discovery of MicroRNAs
1.2. First Implications of miRNAs in Cancer
2. Colon Cancer and miRNAs
2.1. Brief Overview of Colon Cancer
2.2. Early Implications of miRNAs in Colon Cancer
3. Canonical Biogenesis of miRNAs
3.1. Biogenesis in the Nucleus
3.2. Biogenesis in the Cytoplasm
3.3. miRNA-Induced Silencing Complex Formation
| Gene/Protein ID | Location | Function in Biogenesis | Citations |
|---|---|---|---|
| SMAD4 | Nucleus | SMAD4, in complex with SMAD2, is involved in the TGFβ/BMP signaling pathway, and upon activation, they both are able to activate miRNA precursor transcription. | [39,40,41,42] |
| SMAD2 | Nucleus | SMAD4, in complex with SMAD2, is involved in the TGFβ/BMP signaling pathway, and upon activation, they both are able to activate miRNA precursor transcription. SMAD2 also expedites DROSHA processing of primary miRNAs within the nucleus. | [39,42,43,44] |
| DDX17 | Nucleus | DDX17 recruits DROSHA as well as playing a role in the binding of primary miRNAs by DROSHA. | [31,42,43] |
| SRSF3 | Nucleus | SRSF3 binds to CNNC motifs and recruits DROSHA to the basal junction for processing of primary miRNAs. | [28,29] |
| PACT | Cytoplasm | PACT synchronizes precursor miRNA cleavage by DICER1 and contributes to miRISC assembly. | [42,45,46] |
| KHSRP | Cytoplasm | KHSRP promotes the maturation of a subset of miRNAs by binding at their terminal loop. | [42,47] |
| ADAR | Cytoplasm | Adenosine deaminase acting on RNA (ADAR) is a double-stranded RNA-specific enzyme that can act on primary and precursor miRNA stem loops to deaminate an adenosine to an inosine. This can inhibit precursor miRNA cleavage by DICER1. | [42,48,49] |
| LIN28A/B | Cytoplasm | LIN28A/B plays a role in inhibiting DICER1 cleavage by binding to the terminal loop of a subset of precursor miRNAs and recruiting ZCCHC11 or ZCCHC6 that leads to uridylation of precursor miRNAs. | [42,50,51,52] |
| ZCCHC11 (TUT4) and ZCCHC6 (TUT7) | Cytoplasm | Uridylates precursor miRNAs, leading to DICER1 processing inhibition. | [42,51,53] |
| TUDOR-SN | Cytoplasm | TUDOR-SN is a ribonuclease, specific to inosine, containing primary transcripts. | [54] |
| TNRC6A | Cytoplasm | TNRC6A interacts with AGO proteins and plays a role in mRNA degradation. | [42,55,56] |
| PARN | Cytoplasm | PARN is an (A)-specific ribonuclease that degrades miRNAs that have been adenylated by PAPD4 and PAPD5. | [57] |
| PAPD4 and PAPD5 | Cytoplasm | PAPD4 and PAPD5 are nucleotidyl transferases, with adenosyltransferase activity, that lead to miRNA degradation. | [57,58] |
4. Discovery of miRNA Isoforms and Their Classifications
5. Generation of Isoforms through the Biogenesis Pathway
5.1. DROSHA/DICER-Induced Inaccuracies
5.2. Biogenesis Mutations Characterized in Colon Cancers
6. Post-Transcriptional Modifications
6.1. Nucleotidyl Transferase Activity
6.2. Adenosine Deaminase Acting on RNA
6.3. Single-Nucleotide Polymorphisms
7. Functional Importance of IsomiRs
8. Specificity of IsomiRs in Cancer
9. Methods
10. IsomiRs and Colorectal Cancer
10.1. Jiao et al., 2017
10.2. Dokanehiifard et al., 2017
10.3. Wu et al., 2018
10.4. Mjelle et al., 2019
10.5. Nersisyan et al., 2021
10.6. Raigorodskaya et al., 2022
10.7. Lukosevicius et al., 2022
11. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-4. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Reinhart, B.J.; Slack, F.J.; Basson, M.; Pasquinelli, A.E.; Bettinger, J.C.; Rougvie, A.E.; Horvitz, H.R.; Ruvkun, G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 2000, 403, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Chávez Montes, R.A.; De Fátima Rosas-Cárdenas, F.; De Paoli, E.; Accerbi, M.; Rymarquis, L.A.; Mahalingam, G.; Marsch-Martínez, N.; Meyers, B.C.; Green, P.J.; de Folter, S. Sample sequencing of vascular plants demonstrates widespread conservation and divergence of microRNAs. Nat. Commun. 2014, 5, 3722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.-C.; Chan, W.-C.; Hu, L.-Y.; Lai, C.-H.; Hsu, C.-N.; Lin, W.-C. Identification of homologous microRNAs in 56 animal genomes. Genomics 2014, 96, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 121–134. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [Green Version]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [Green Version]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global burden of colorectal cancer in 2020 and 2040: Incidence and mortality estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Akimoto, N.; Ugai, T.; Zhong, R.; Hamada, T.; Fujiyoshi, K.; Giannakis, M.; Wu, K.; Cao, Y.; Ng, K.; Ogino, S. Rising incidence of early-onset colorectal cancer—A call to action. Nat. Rev. Clin. Oncol. 2021, 18, 230–243. [Google Scholar] [CrossRef] [PubMed]
- Al-Akhrass, H.; Christou, N. The Clinical Assessment of MicroRNA Diagnostic, Prognostic, and Theranostic Value in Colorectal Cancer. Cancers 2021, 13, 2916. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vychytilova-Faltejskova, P.; Radova, L.; Sachlova, M.; Kosarova, Z.; Slaba, K.; Fabian, P.; Grolich, T.; Prochazka, V.; Kala, Z.; Svoboda, M.; et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinog. 2016, 37, 941–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- To, K.K.; Tong, C.W.; Wu, M.; Cho, W.C. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J. Gastroenterol. 2018, 24, 2949–2973. [Google Scholar] [CrossRef] [PubMed]
- Michael, M.Z.; O’ Connor, S.M.; van Holst Pellekaan, N.G.; Young, G.P.; James, R.J. Reduced Accumulation of Specific MicroRNAs in Colorectal Neoplasia. Mol. Cancer Res. 2003, 1, 882–891. [Google Scholar]
- Pidíkova, P.; Reis, R.; Herichova, I. miRNA Clusters with Down-Regulated Expression in Human Colorectal Cancer and Their Regulation. Int. J. Mol. Sci. 2020, 21, 4633. [Google Scholar] [CrossRef]
- Pidíková, P.; Herichová, I. miRNA Clusters with Up-Regulated Expression in Colorectal Cancer. Cancers 2021, 13, 2979. [Google Scholar] [CrossRef]
- Yang, Z.; Wu, L.; Wang, A.; Tang, W.; Zhao, Y.; Zhao, H.; Teschendorff, A.E. dbDEMC 2.0: Updated database of differentially expressed miRNAs in human cancers. Nucleic Acids Res. 2017, 45, D812–D818. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Zhou, T.; Chen, Q. Exploring the expanding universe of small RNAs. Nat. Cell Biol. 2022, 24, 415–423. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]
- Stavast, C.J.; Erkeland, S.J. The Non-Canonical Aspects of MicroRNAs: Many Roads to Gene Regulation. Cells 2019, 8, 1465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.-H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef] [Green Version]
- Cai, X.; Hagedorn, C.H.; Cullen, B.R. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA 2004, 10, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Auyeung, V.C.; Ulitsky, I.; McGeary, S.E.; Bartel, D.P. Beyond Secondary Structure: Primary-Sequence Determinants License Pri-miRNA Hairpins for Processing. Cell 2013, 152, 844–858. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Nguyen, T.D.; Li, S.; Nguyen, T.A. SRSF3 recruits DROSHA to the basal junction of primary microRNAs. RNA 2018, 24, 892–898. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Lee, Y.; Yeom, K.-H.; Kim, Y.-K.; Jin, H.; Kim, V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004, 18, 3016–3027. [Google Scholar] [CrossRef] [Green Version]
- Gregory, R.I.; Yan, K.-P.; Amuthan, G.; Chendrimada, T.; Doratotaj, B.; Cooch, N.; Shiekhattar, R. The Microprocessor complex mediates the genesis of microRNAs. Nature 2004, 432, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.J.; Plasterk, R.H.A.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [Green Version]
- Park, J.-E.; Heo, I.; Tian, Y.; Simanshu, D.K.; Chang, H.; Jee, D.; Patel, D.J.; Kim, V.N. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 2011, 475, 201–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Kolb, F.A.; Jaskiewicz, L.; Westhof, E.; Filipowicz, W. Single Processing Center Models for Human Dicer and Bacterial RNase III. Cell 2004, 118, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Kretov, D.A.; Walawalkar, I.A.; Mora-Martin, A.; Shafik, A.M.; Moxon, S.; Cifuentes, D. Ago2-Dependent Processing Allows miR-451 to Evade the Global MicroRNA Turnover Elicited during Erythropoiesis. Mol. Cell 2020, 78, 317–328.e6. [Google Scholar] [CrossRef] [PubMed]
- Yoda, M.; Kawamata, T.; Paroo, Z.; Ye, X.; Iwasaki, S.; Liu, Q.; Tomari, Y. ATP-dependent human RISC assembly pathways. Nat. Struct. Mol. Biol. 2010, 17, 17–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medley, J.C.; Panzade, G.; Zinovyeva, A.Y. microRNA strand selection: Unwinding the rules. Wiley Interdiscip. Rev. RNA 2021, 12, e1627. [Google Scholar] [CrossRef]
- Davis, B.N.; Hilyard, A.C.; Lagna, G.; Hata, A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature 2008, 454, 56–61. [Google Scholar] [CrossRef] [Green Version]
- Blahna, M.T.; Hata, A. Smad-mediated regulation of microRNA biosynthesis. FEBS Lett. 2012, 586, 1906–1912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, S.; He, X.; Ding, J.; Liang, L.; Zhao, Y.; Zhang, Z.; Yao, X.; Pan, Z.; Zhang, P.; Li, J.; et al. Upregulation of miR-23a∼27a∼24 decreases transforming growth factor-beta-induced tumor-suppressive activities in human hepatocellular carcinoma cells. Int. J. Cancer 2008, 123, 972–978. [Google Scholar] [CrossRef]
- Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; Nawrocka, P.M.; Kozlowski, P. A pan-cancer atlas of somatic mutations in miRNA biogenesis genes. Nucleic Acids Res. 2021, 49, 601–620. [Google Scholar] [CrossRef]
- Davis-Dusenbery, B.; Hilyard, A.C.; Nguyen, P.H.; Lagna, G.; Hata, A. Smad Proteins Bind a Conserved RNA Sequence to Promote MicroRNA Maturation by Drosha. Mol. Cell 2010, 39, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Warner, D.R.; Bhattacherjee, V.; Yin, X.; Singh, S.; Mukhopadhyay, P.; Pisano, M.M.; Greene, R.M. Functional interaction between Smad, CREB binding protein, and p68 RNA helicase. Biochem. Biophys. Res. Commun. 2004, 324, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Zhou, K.; Smith, A.M.; Noland, C.L.; Doudna, J.A. Differential roles of human Dicer-binding proteins TRBP and PACT in small RNA processing. Nucleic Acids Res. 2013, 41, 6568–6576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.; Hur, I.; Park, S.-Y.; Kim, Y.K.; Suh, M.R.; Kim, V.N. The role of PACT in the RNA silencing pathway. EMBO J. 2006, 25, 522–532. [Google Scholar] [CrossRef]
- Trabucchi, M.; Briata, P.; Filipowicz, W.; Ramos, A.; Gherzi, R.; Rosenfeld, M.G. KSRP promotes the maturation of a group of miRNA precursors. Adv. Exp. Med. Biol. 2010, 700, 36–42. [Google Scholar]
- Tomaselli, S.; Bonamassa, B.; Alisi, A.; Nobili, V.; Locatelli, F.; Gallo, A. ADAR Enzyme and miRNA Story: A Nucleotide that Can Make the Difference. Int. J. Mol. Sci. 2013, 14, 22796–22816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.; Chendrimada, T.P.; Wang, Q.; Higuchi, M.; Seeburg, P.H.; Shiekhattar, R.; Nishikura, K. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006, 13, 13–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nowak, J.S.; Choudhury, N.R.; De Lima Alves, F.; Rappsilber, J.; Michlewski, G. Lin28a regulates neuronal differentiation and controls miR-9 production. Nat. Commun. 2014, 5, 3687. [Google Scholar] [CrossRef] [Green Version]
- Heo, I.; Joo, C.; Kim, Y.-K.; Ha, M.; Yoon, M.-J.; Cho, J.; Yeom, K.-H.; Han, J.; Kim, V.N. TUT4 in Concert with Lin28 Suppresses MicroRNA Biogenesis through Pre-MicroRNA Uridylation. Cell 2009, 138, 696–708. [Google Scholar] [CrossRef] [Green Version]
- Viswanathan, S.R.; Daley, G.Q.; Gregory, R.I. Selective Blockade of MicroRNA Processing by Lin28. Science 2008, 320, 97–100. [Google Scholar] [CrossRef] [Green Version]
- Thornton, J.E.; Du, P.; Jing, L.; Sjekloca, L.; Lin, S.; Grossi, E.; Sliz, P.; Zon, L.I.; Gregory, R.I. Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Res. 2014, 42, 11777–11791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.-L.; Yang, W.-Z.; Chen, Y.-P.; Yuan, H.S. Structural and functional insights into human Tudor-SN, a key component linking RNA interference and editing. Nucleic Acids Res. 2008, 36, 3579–3589. [Google Scholar] [CrossRef] [Green Version]
- Jakymiw, A.; Lian, S.; Eystathioy, T.; Li, S.; Satoh, M.; Hamel, J.C.; Fritzler, M.J.; Chan, E.K. Disruption of GW bodies impairs mammalian RNA interference. Nat. Cell Biol. 2005, 7, 1267–1274. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Rivas, F.V.; Wohlschlegel, J.; Yates, J.R., III; Parker, R.; Hannon, G.J. A role for the P-body component GW182 in microRNA function. Nat. Cell Biol. 2005, 7, 1261–1266. [Google Scholar] [CrossRef]
- Boele, J.; Persson, H.; Shin, J.W.; Ishizu, Y.; Newie, I.S.; Søkilde, R.; Hawkins, S.M.; Coarfa, C.; Ikeda, K.; Takayama, K.-I.; et al. PAPD5-mediated 3′ adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proc. Natl. Acad. Sci. USA 2014, 111, 11467–11472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, T.; Sakaguchi, Y.; Miyauchi, K.; Suzuki, T.; Kashiwabara, S.-I.; Baba, T.; Suzuki, T. Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD-2. Genes Dev. 2009, 23, 433–438. [Google Scholar] [CrossRef] [Green Version]
- Bofill-De Ros, X.; Hong, Z.; Birkenfeld, B.; Alamo-Ortiz, S.; Yang, A.; Dai, L.; Gu, S. Flexible pri-miRNA structures enable tunable production of 5′ isomiRs. RNA Biol. 2022, 19, 279–289. [Google Scholar] [CrossRef]
- Creugny, A.; Fender, A.; Pfeffer, S. Regulation of primary microRNA processing. FEBS Lett. 2018, 592, 1980–1996. [Google Scholar] [CrossRef] [PubMed]
- Michlewski, G.; Cáceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shkurnikov, M.Y.; Nersisyan, S.A.; Osepyan, A.S.; Maltseva, D.V.; Knyazev, E.N. Differences in the Drosha and Dicer Cleavage Profiles in Colorectal Cancer and Normal Colon Tissue Samples. Dokl. Biochem. Biophys. 2020, 493, 208–210. [Google Scholar] [CrossRef]
- Zhiyanov, A.; Nersisyan, S.; Tonevitsky, A. Hairpin sequence and structure is associated with features of isomiR biogenesis. RNA Biol. 2021, 18, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Yeo, J.; Lee, J.H.; Cho, J.; Seo, D.; Kim, J.-S.; Kim, V.N. Deletion of Human tarbp2 Reveals Cellular MicroRNA Targets and Cell-Cycle Function of TRBP. Cell Rep. 2014, 9, 1061–1074. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.Y.; Doudna, J.A. TRBP alters human precursor microRNA processing in vitro. RNA 2012, 18, 2012–2019. [Google Scholar] [CrossRef] [Green Version]
- Wilson, R.C.; Tambe, A.; Kidwell, M.A.; Noland, C.L.; Schneider, C.P.; Doudna, J.A. Dicer-TRBP Complex Formation Ensures Accurate Mammalian MicroRNA Biogenesis. Mol. Cell 2015, 57, 397–407. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.-W.; Pan, J.-J.; Hu, J.-F.; Zhang, J.-Q.; Huang, L.; Huang, Y.; Liao, C.-Y.; Yang, C.; Chen, Z.-W.; Wang, Y.-D.; et al. SRSF3-mediated regulation of N6-methyladenosine modification-related lncRNA ANRIL splicing promotes resistance of pancreatic cancer to gemcitabine. Cell Rep. 2022, 39, 110813. [Google Scholar] [CrossRef] [PubMed]
- Vychytilova-Faltejskova, P.; Svobodova Kovarikova, A.; Grolich, T.; Prochazka, V.; Slaba, K.; Machackova, T.; Halamkova, J.; Svoboda, M.; Kala, Z.; Kiss, I.; et al. MicroRNA Biogenesis Pathway Genes Are Deregulated in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papachristou, D.J.; Korpetinou, A.; Giannopoulou, E.; Antonacopoulou, A.G.; Papadaki, H.; Grivas, P.; Scopa, C.D.; Kalofonos, H.P. Expression of the ribonucleases Drosha, Dicer, and Ago2 in colorectal carcinomas. Virchows Arch. 2011, 459, 431–440. [Google Scholar] [CrossRef]
- Faber, C.; Horst, D.; Hlubek, F.; Kirchner, T. Overexpression of Dicer predicts poor survival in colorectal cancer. Eur. J. Cancer 2011, 47, 1414–1419. [Google Scholar] [CrossRef]
- Stratmann, J.; Wang, C.-J.; Gnosa, S.; Wallin, A.; Hinselwood, D.; Sun, X.-F.; Zhang, H. Dicer and miRNA in relation to clinicopathological variables in colorectal cancer patients. BMC Cancer 2011, 11, 345. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Song, M.L.; Min, H.; Hwang, I.; Baek, S.K.; Kwon, T.K.; Park, J.-W. miRNA biogenesis-associated RNase III nucleases Drosha and Dicer are upregulated in colorectal adenocarcinoma. Oncol. Lett. 2017, 14, 4379–4383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faggad, A.; Kasajima, A.; Weichert, W.; Stenzinger, A.; Elwali, N.E.; Dietel, M.; Denkert, C. Down-regulation of the microRNA processing enzyme Dicer is a prognostic factor in human colorectal cancer. Histopathology 2012, 61, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Sheu-Gruttadauria, J.; Pawlica, P.; Klum, S.M.; Wang, S.; Yario, T.A.; Schirle Oakdale, N.T.; Steitz, J.A.; MacRae, I.J. Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell 2019, 75, 1243–1255.e7. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Shao, T.-J.; Bofill-De Ros, X.; Lian, C.; Villanueva, P.; Dai, L.; Gu, S. AGO-bound mature miRNAs are oligouridylated by TUTs and subsequently degraded by DIS3L2. Nat. Commun. 2020, 11, 2765. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Sheng, P.; Li, T.; Fields, C.J.; Hiers, N.M.; Wang, Y.; Li, J.; Guardia, C.M.; Licht, J.D.; Xie, M. Widespread microRNA degradation elements in target mRNAs can assist the encoded proteins. Genes Dev. 2021, 35, 1595–1609. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Bofill-De Ros, X.; Stanton, R.; Shao, T.-J.; Villanueva, P.; Gu, S. TENT2, TUT4, and TUT7 selectively regulate miRNA sequence and abundance. Nat. Commun. 2022, 13, 5260. [Google Scholar] [CrossRef]
- Han, J.; Mendell, J.T. MicroRNA turnover: A tale of tailing, trimming, and targets. Trends Biochem. Sci. 2022, 48, 26–39. [Google Scholar] [CrossRef]
- Kawahara, Y.; Zinshteyn, B.; Chendrimada, T.P.; Shiekhattar, R.; Nishikura, K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer–TRBP complex. EMBO Rep. 2007, 8, 763–769. [Google Scholar] [CrossRef] [Green Version]
- Tsuchiya, N.; Ochiai, M.; Nakashima, K.; Ubagai, T.; Sugimura, T.; Nakagama, H. SND1, a Component of RNA-Induced Silencing Complex, Is Up-regulated in Human Colon Cancers and Implicated in Early Stage Colon Carcinogenesis. Cancer Res 2007, 67, 9568–9576. [Google Scholar] [CrossRef] [Green Version]
- Kawakara, Y.; Zinshteyn, B.; Sethupathy, P.; Iizasa, H.; Hatzigeorgiou, A.G.; Nishikura, K. Redirection of Silencing Targets by Adenosine-to-Inosine editing of miRNAs. Science 2007, 315, 1133–1137. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Song, Y.; Shi, X.; Liu, J.; Xiong, S.; Chen, W.; Fu, Q.; Huang, Z.; Gu, N.; Zhang, R. The landscape of miRNA editing in animals and its impact on miRNA biogenesis and targeting. Genome Res. 2018, 28, 132–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, L.; Song, Y.; Chan, T.H.M.; Yang, H.; Lin, C.H.; Tay, D.J.T.; Hong, H.; Tang, S.J.; Tan, K.T.; Huang, X.X.; et al. An RNA editing/dsRNA binding-independent gene regulatory mechanism of ADARs and its clinical implication in cancer. Nucleic Acids Res. 2017, 45, 10436–10451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Lin, D.-H.; Xu, J.-P.; Chen, W.-X.; Zheng, S.-J.; Song, L. Genotype GG of rs895819 Functional Polymorphism Within miR-27a Might Increase Genetic Susceptibility to Colorectal Cancer in Han Chinese Population. J. Clin. Lab. Anal. 2016, 30, 351–355. [Google Scholar] [CrossRef]
- Hezova, R.; Kovarikova, A.; Bienertova-Vasku, J.; Sachlova, M.; Redova, M.; Vasku, A.; Svoboda, M.; Radova, L.; Kiss, I.; Vyzula, R.; et al. Evaluation of SNPs in miR-196-a2, miR-27a and miR-146a as risk factors of colorectal cancer. World J. Gastroenterol. 2012, 18, 2827–2831. [Google Scholar] [CrossRef]
- Bian, Q.; Chen, J.-J.; Gu, J.-P.; Xu, J. Association between pre-miR-27a functional polymorphism and risk of colorectal cancer in north Chinese Han population. OncoTargets Ther. 2015, 8, 3003–3007. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Sun, X.; Wang, Y.; Liu, X.; Xuan, Y.; Hu, S. Association between miR-27a genetic variants and susceptibility to colorectal cancer. Diagn. Pathol. 2014, 9, 146. [Google Scholar] [CrossRef] [Green Version]
- Kupcinskas, J.; Bruzaite, I.; Juzenas, S.; Gyvyte, U.; Jonaitis, L.; Kiudelis, G.; Skieceviciene, J.; Leja, M.; Pauzas, H.; Tamelis, A.; et al. Lack of association between miR-27a, miR-146a, miR-196a-2, miR-492 and miR-608 gene polymorphisms and colorectal cancer. Sci. Rep. 2014, 4, srep05993. [Google Scholar] [CrossRef]
- Cao, Y.; Hu, J.; Fang, Y.; Chen, Q.; Li, H. Association between a functional variant in microRNA-27a and susceptibility to colorectal cancer in a Chinese Han population. Genet. Mol. Res. 2014, 13, 7420–7427. [Google Scholar] [CrossRef]
- Dai, J.; Chen, Y.; Gong, Y.; Gu, D.; Chen, J. Association of microRNA-27a rs895819 polymorphism with the risk of cancer: An updated meta-analysis. Gene 2020, 728, 144185. [Google Scholar] [CrossRef]
- Bofill-De Ros, X.; Yang, A.; Gu, S. IsomiRs: Expanding the miRNA repression toolbox beyond the seed. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2020, 1863, 194373. [Google Scholar] [CrossRef]
- Zhou, L.; Yu, M.; Lim, T.; Kaur, P.; Saj, A.; Bortolamiol-Becet, D.; Gopal, V.; Tucker-Kellog, G.; Okamura, K. Importance of miRNA stability and alternative primary miRNA isoforms in gene regulation during Drosophila development. eLife 2018, 7, e38389. [Google Scholar] [CrossRef]
- Broughton, J.P.; Lovci, M.T.; Huang, J.L.; Yeo, G.W.; Pasquinelli, A.E. Pairing beyond the Seed Supports MicroRNA Targeting Specificity. Mol. Cell 2016, 64, 320–333. [Google Scholar] [CrossRef] [Green Version]
- Moore, M.J.; Scheel, T.K.H.; Luna, J.M.; Park, C.Y.; Fak, J.J.; Nishiuchi, E.; Rice, C.M.; Darnell, R.B. miRNA–target chimeras reveal miRNA 3′-end pairing as a major determinant of Argonaute target specificity. Nat. Commun. 2015, 6, 8864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burroughs, A.M.; Ando, Y.; De Hoon, M.J.L.; Tomaru, Y.; Suzuki, H.; Hayashizaki, Y.; Daub, C. Deep-sequencing of human Argonaute-associated small RNAs provides insight into miRNA sorting and reveals Argonaute association with RNA fragments of diverse origin. RNA Biol. 2011, 8, 158–177. [Google Scholar] [CrossRef] [PubMed]
- Telonis, A.G.; Magee, R.; Loher, P.; Chervoneva, I.; Londin, E.; Rigoutsos, I. Knowledge about the presence or absence of miRNA isoforms (isomiRs) can successfully discriminate amongst 32 TCGA cancer types. Nucleic Acids Res. 2017, 45, 2973–2985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Zheng, Z.; Chen, P.; Wu, M. Tumor classification and biomarker discovery based on the 5′isomiR expression level. BMC Cancer 2019, 19, 127. [Google Scholar] [CrossRef]
- Jiao, W.; Leng, X.; Zhou, Q.; Wu, Y.; Sun, L.; Tan, Y.; Ni, H.; Dong, X.; Shen, T.; Liu, Y.; et al. Different miR-21-3p isoforms and their different features in colorectal cancer. Int. J. Cancer 2017, 141, 2103–2111. [Google Scholar] [CrossRef] [Green Version]
- Dokanehiifard, S.; Yasari, A.; Najafi, H.; Jafarzadeh, M.; Nikkhah, M.; Mowla, S.J.; Soltani, B.M. A novel microRNA located in the TrkC gene regulates the Wnt signaling pathway and is differentially expressed in colorectal cancer specimens. J. Biol. Chem. 2017, 292, 7566–7577. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.W.; Evans, J.M.; Huang, S.; Mahoney, D.W.; Dukek, B.A.; Taylor, W.R.; Yab, T.C.; Smyrk, T.C.; Jen, J.; Kisiel, J.B.; et al. A Comprehensive Approach to Sequence-oriented IsomiR annotation (CASMIR): Demonstration with IsomiR profiling in colorectal neoplasia. BMC Genom. 2018, 19, 401. [Google Scholar] [CrossRef]
- Kabekkodu, S.P.; Shukla, V.; Varghese, V.K.; D’Souza, J.; Chakrabarty, S.; Satyamoorthy, K. Clustered miRNAs and their role in biological functions and diseases. Biol. Rev. 2018, 93, 1955–1986. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Gupta, A.; Krawczyk, J.; Gupta, S. The miR-17-92 cluster: Yin and Yang in human cancers. Cancer Treat. Res. Commun. 2022, 33, 100647. [Google Scholar] [CrossRef] [PubMed]
- Fang, L.-L.; Wang, X.-H.; Sun, B.-F.; Zhang, X.-D.; Zhu, X.-H.; Yu, Z.-J.; Luo, H. Expression, regulation and mechanism of action of the miR-17-92 cluster in tumor cells (Review). Int. J. Mol. Med. 2017, 40, 1624–1630. [Google Scholar] [CrossRef] [Green Version]
- Mjelle, R.; Sjursen, W.; Thommesen, L.; Sætrom, P.; Hofsli, E. Small RNA expression from viruses, bacteria and human miRNAs in colon cancer tissue and its association with microsatellite instability and tumor location. BMC Cancer 2019, 19, 161. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.-J.; Yao, Y.; Cai, X.-Y.; Fang, G.-Y. Emerging Role of MiR-192-5p in Human Diseases. Front. Pharmacol. 2021, 12, 614068. [Google Scholar] [CrossRef] [PubMed]
- Nersisyan, S.; Novosad, V.; Engibaryan, N.; Ushkaryov, Y.; Nikulin, S.; Tonevitsky, A. ECM–Receptor Regulatory Network and Its Prognostic Role in Colorectal Cancer. Front. Genet. 2021, 12, 782699. [Google Scholar] [CrossRef] [PubMed]
- Raigorodskaya, M.P.; Zhiyanov, A.P.; Averinskaya, D.A.; Tonevitsky, E.A. Changes in the Expression of miRNA Isoforms and Their Targets in HT-29 Cells after Hypoxic Exposure. Bull. Exp. Biol. Med. 2022, 173, 123–127. [Google Scholar] [CrossRef]
- Lukosevicius, R.; Juzenas, S.; Salteniene, V.; Kulokiene, U.; Arstikyte, J.; Hemmrich-Stanisak, G.; Franke, A.; Link, A.; Ruzgys, P.; Satkauskas, S.; et al. miRNome Profiling and Functional Analysis Reveal Involvement of hsa-miR-1246 in Colon Adenoma-Carcinoma Transition by Targeting AXIN2 and CFTR. Int. J. Mol. Sci. 2022, 23, 2107. [Google Scholar] [CrossRef]
- Jenike, A.E.; Halushka, M.K. miR-21: A non-specific biomarker of all maladies. Biomark. Res. 2021, 9, 18. [Google Scholar] [CrossRef]
- Dong, Y.; Yu, J.; Ng, S.S. MicroRNA dysregulation as a prognostic biomarker in colorectal cancer. Cancer Manag. Res. 2014, 6, 405–422. [Google Scholar] [CrossRef] [Green Version]
- Bautista-Sánchez, D.; Arriaga-Canon, C.; Pedroza-Torres, A.; De La Rosa-Velázquez, I.A.; González-Barrios, R.; Contreras-Espinosa, L.; Montiel-Manríquez, R.; Castro-Hernández, C.; Fragoso-Ontiveros, V.; Álvarez-Gómez, R.M.; et al. The promising role of miR-21 as a cancer biomarker and its importance in RNA-Based therapeutics. Mol. Ther. Nucleic. Acids. 2020, 20, 409–420. [Google Scholar] [CrossRef]
- Zhao, H.; Chen, J.; Chen, J.; Kong, X.; Zhu, H.; Zhang, Y.; Dong, H.; Wang, J.; Ren, Q.; Wang, Q.; et al. miR-192/215-5p act as tumor suppressors and link Crohn’s disease and colorectal cancer by targeting common metabolic pathways: An integrated informatics analysis and experimental study. J. Cell. Physiol. 2019, 234, 21060–21075. [Google Scholar] [CrossRef] [PubMed]
- Chiang, Y.; Song, Y.; Wang, Z.; Liu, Z.; Gao, P.; Liang, J.; Zhu, J.; Xing, C.; Xu, H. microRNA-192, -194 and -215 are frequently downregulated in colorectal cancer. Exp. Ther. Med. 2012, 3, 560–566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsikitis, V.L.; White, I.; Mori, M.; Potter, A.; Bhattcharyya, A.; Hamilton, S.R.; Buckmeier, J.; Lance, P.; Thompson, P. Differential expression of microRNA-320a, -145, and -192 along the continuum of normal mucosa to high-grade dysplastic adenomas of the colorectum. Am. J. Surg. 2014, 207, 717–722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zelli, V.; Compagnoni, C.; Capelli, R.; Corrente, A.; Cornice, J.; Vecchiotti, D.; Di Padova, M.; Zazzeroni, F.; Alesse, E.; Tessitore, A. Emerging Role of isomiRs in Cancer: State of the Art and Recent Advances. Genes 2021, 12, 1447. [Google Scholar] [CrossRef] [PubMed]


| Adenomatous Polyps vs. Healthy Controls | |||
| Gene | Base Mean | Log2 Fold Change | Adjusted p-Value |
| miR-27a-3p | 9614.9 | 0.343 | 0.006 |
| miR-27a-3p.iso.t5:0.seed:0.t3:cgc.ad:GG.mm:0 | 64.7 | 0.681 | 0.004 |
| miR-27a-3p.iso.t5:0.seed:0.t3:0.ad:AAT.mm:0 | 1.4 | 1.293 | 0.028 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:TAT.mm:0 | 3.6 | 0.824 | 0.032 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:0.mm:0 | 5347.8 | 0.336 | 0.038 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:AAC.mm:0 | 5.1 | 0.607 | 0.044 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:GAT.mm:0 | 0.9 | 1.259 | 0.045 |
| miR-27a-3p.iso.t5:0.seed:0.t3:cgc.ad:0.mm:0 | 505.1 | 0.813 | <0.001 |
| miR-27a-3p.iso.t5:0.seed:0.t3:gc.ad:0.mm:0 | 1192.9 | 0.554 | <0.001 |
| Colorectal Cancer vs. Adenomatous Polyps | |||
| Gene | Base Mean | Log2 Fold Change | Adjusted p-Value |
| hsa-miR-27a-3p | 9614.9 | 0.343 | 0.006 |
| miR-27a-3p.iso.t5:0.seed:0.t3:cgc.ad:0.mm:0 | 505.1 | −0.675 | <0.001 |
| miR-27a-3p.iso.t5:0.seed:0.t3:cgc.ad:GG.mm:0 | 64.7 | −0.671 | 0.010 |
| miR-27a-3p.iso.t5:0.seed:0.t3:0.ad:A.mm:0 | 62.0 | 0.578 | 0.010 |
| miR-27a-3p.iso.t5:0.seed:0.t3:C.ad:0.mm:0 | 7.2 | 0.668 | 0.017 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:TT.mm:0 | 119.3 | 0.701 | 0.017 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:TA.mm:0 | 5.3 | 0.705 | 0.025 |
| miR-27a-3p.ref.t5:0.seed:0.t3:0.ad:0.mm:0 | 861.6 | 0.473 | 0.032 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:0.mm:0 | 5347.8 | 0.367 | 0.035 |
| miR-27a-3p.iso.t5:0.seed:0.t3:gc.ad:TTT.mm:0 | 3.2 | 0.815 | 0.038 |
| miR-27a-3p.iso.t5:0.seed:0.t3:gc.ad:A.mm:0 | 20.1 | 1.003 | <0.001 |
| Colorectal Cancer vs. Healthy Controls | |||
| Gene | Base Mean | Log2 Fold Change | Adjusted p-Value |
| hsa-miR-27a-3p | 9614.9 | 0.343 | 0.006 |
| miR-27a-3p.iso.t5:0.seed:0.t3:gc.ad:0.mm:0 | 1192.9 | 0.793 | <0.001 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:0.mm:0 | 5347.8 | 0.704 | <0.001 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:T.mm:0 | 223.6 | 0.710 | <0.001 |
| miR-27a-3p.ref.t5:0.seed:0.t3:0.ad:0.mm:0 | 861.61 | 0.652 | <0.001 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:TC.mm:0 | 2.0 | 1.190 | 0.002 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:AAC.mm:0 | 5.1 | 0.738 | 0.005 |
| miR-27a-3p.iso.t5:0.seed:0.t3:0.ad:A.mm:0 | 62.0 | 0.568 | 0.007 |
| miR-27a-3p.iso.t5:0.seed:0.t3:0.ad:AAT.mm:0 | 1.4 | 1.476 | 0.007 |
| miR-27a-3p.iso.t5:0.seed:0.t3:gc.ad:A.mm:0 | 20.1 | 0.566 | 0.0218 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:ATT.mm:0 | 1.7 | 1.058 | 0.025 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:AC.mm:0 | 1.7 | 1.004 | 0.029 |
| miR-27a-3p.iso.t5:0.seed:0.t3:0.ad:G.mm:0 | 17.6 | 0.388 | 0.047 |
| miR-27a-3p.iso.t5:0.seed:0.t3:c.ad:TCT.mm:0 | 1.7 | 0.851 | 0.048 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lausten, M.A.; Boman, B.M. A Review of IsomiRs in Colorectal Cancer. Non-Coding RNA 2023, 9, 34. https://doi.org/10.3390/ncrna9030034
Lausten MA, Boman BM. A Review of IsomiRs in Colorectal Cancer. Non-Coding RNA. 2023; 9(3):34. https://doi.org/10.3390/ncrna9030034
Chicago/Turabian StyleLausten, Molly A., and Bruce M. Boman. 2023. "A Review of IsomiRs in Colorectal Cancer" Non-Coding RNA 9, no. 3: 34. https://doi.org/10.3390/ncrna9030034
APA StyleLausten, M. A., & Boman, B. M. (2023). A Review of IsomiRs in Colorectal Cancer. Non-Coding RNA, 9(3), 34. https://doi.org/10.3390/ncrna9030034