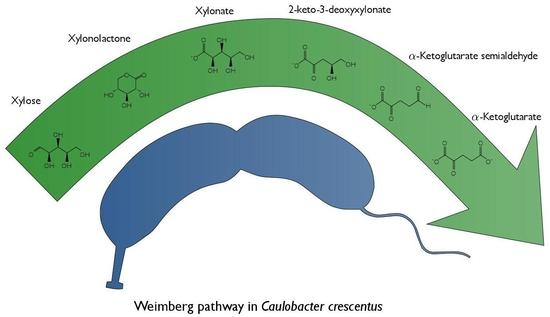
Characterization of the Weimberg Pathway in Caulobacter crescentus
Abstract
:1. Introduction
2. Results
2.1. Growth in Shake Flasks
2.2. Enzyme Activity
2.3. Characterization in Bioreactors
3. Discussion
4. Materials and Methods
4.1. Microorganism
4.2. Medium Composition
4.3. Shake-Flask Cultivation
4.4. Bioreactor Cultivation
4.5. Biomass Measurements
4.6. Analyses of Sugars and Organic Acids
4.7. Enzymatic Assays
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CDW | Cell Dry Weight |
| DOT | Dissolved Oxygen Tension |
| HPLC | High Performance Liquid Chromatography |
| LG | Lower Glycolysis |
| NADH/NAD+ | Nicotinamide Adenine Dinucleotide (Reduced/Oxidized) |
| OD | Optical Density |
| PDH | Pyruvate dehydrogenase |
| PPP | Pentose Phosphate Pathway |
| TCA-Cycle | Tricarboxylic Acid Cycle |
| UHPLC | Ultra High Performance Liquid Chromatography |
| XDH | Xylulose Dehydrogenase |
| XI | Xylose Isomerase |
| XR | Xylose Reductase |
References
- Henrici, A.T.; Johnson, D.E. Studies of Freshwater Bacteria: II. Stalked Bacteria, a New Order of Schizomycetes. J. Bacteriol. 1935, 30, 61–93. [Google Scholar] [PubMed]
- Skerker, J.M.; Laub, M.T. Cell-cycle progression and the generation of asymmetry in Caulobacter crescentus. Nat. Rev. Microbiol. 2004, 2, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Curtis, P.D.; Brun, Y.V. Getting in the loop: Regulation of development in Caulobacter crescentus. Microbiol. Mol. Biol. Rev. MMBR 2010, 74, 13–41. [Google Scholar] [CrossRef] [PubMed]
- Tsokos, C.G.; Laub, M.T. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr. Opin. Microbiol. 2012, 15, 744–750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nierman, W.C.; Feldblyum, T.V.; Laub, M.T.; Paulsen, I.T.; Nelson, K.E.; Eisen, J.; Heidelberg, J.F.; Alley, M.R.K.; Ohta, N.; Maddock, J.R.; et al. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. USA 2001, 98, 4136–4141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hottes, A.K.; Meewan, M.; Yang, D.; Arana, N.; Romero, P.; McAdams, H.H.; Stephens, C. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 2004, 186, 1448–1461. [Google Scholar] [CrossRef] [PubMed]
- Arellano, B.H.; Ortiz, J.D.; Manzano, J.; Chen, J.C. Identification of a dehydrogenase required for lactose metabolism in caulobacter crescentus. Appl. Environ. Microbiol. 2010, 76, 3004–3014. [Google Scholar] [CrossRef] [PubMed]
- Zalatan, F.; Black, P. Characterization of long-chain fatty acid uptake in Caulobacter crescentus. Arch. Microbiol. 2011, 193, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Presley, G.N.; Payea, M.J.; Hurst, L.R.; Egan, A.E.; Martin, B.S.; Periyannan, G.R. Extracellular gluco-oligosaccharide degradation by Caulobacter crescentus. Microbiology 2014, 160, 635–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wichelecki, D.J.; Graff, D.C.; Al-Obaidi, N.; Almo, S.C.; Gerlt, J.A. Identification of the in Vivo Function of the High-Efficiency d-Mannonate Dehydratase in Caulobacter crescentus NA1000 from the Enolase Superfamily. Biochemistry 2014, 53, 4087–4089. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C.; Christen, B.; Fuchs, T.; Sundaram, V.; Watanabe, K.; Jenal, U. Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 2007, 189, 2181–2185. [Google Scholar] [CrossRef] [PubMed]
- Weimberg, R. Pentose oxidation by Pseudomonas fragi. J. Biol. Chem. 1961, 236, 629–635. [Google Scholar] [PubMed]
- Radek, A.; Krumbach, K.; Gätgens, J.; Wendisch, V.F.; Wiechert, W.; Bott, M.; Noack, S.; Marienhagen, J. Engineering of Corynebacterium glutamicum for minimized carbon loss during utilization of d-xylose containing substrates. J. Biotechnol. 2014, 192, 156–160. [Google Scholar] [CrossRef] [PubMed]
- Karhumaa, K.; Garcia Sanchez, R.; Hahn-Hägerdal, B.; Gorwa-Grauslund, M.F. Comparison of the xylose reductase-xylitol dehydrogenase and the xylose isomerase pathways for xylose fermentation by recombinant Saccharomyces cerevisiae. Microb. Cell Fact. 2007, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Álvarez, C.; Reyes-Sosa, F.M.; Díez, B. Enzymatic hydrolysis of biomass from wood. Microb. Biotechnol. 2016, 9, 149–156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meijnen, J.P.; De Winde, J.H.; Ruijssenaars, H.J. Establishment of oxidative d-xylose metabolism in Pseudomonas putida S12. Appl. Environ. Microbiol. 2009, 75, 2784–2791. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Valdehuesa, K.N.G.; Nisola, G.M.; Ramos, K.R.M.; Chung, W.J. High yield production of d-xylonic acid from d-xylose using engineered Escherichia coli. Bioresour. Technol. 2012, 115, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xian, M.; Zou, H.; Zhang, H. Metabolic Engineering of Escherichia coli for the Production of Xylonate. PLoS ONE 2013, 8, e67305. [Google Scholar] [CrossRef] [PubMed]
- Toivari, M.H.; Ruohonen, L.; Richard, P.; Penttilä, M.; Wiebe, M.G. Saccharomyces cerevisiae engineered to produce d-xylonate. Appl. Microbiol. Biotechnol. 2010, 88, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Kohler, C.; Lourenço, R.F.; Bernhardt, J.; Albrecht, D.; Schüler, J.; Hecker, M.; Gomes, S.L. A comprehensive genomic, transcriptomic and proteomic analysis of a hyperosmotic stress sensitive α-proteobacterium. BMC Microbiol. 2015, 15, 71. [Google Scholar] [CrossRef] [PubMed]
- Ely, B. Genetics of Caulobacter crescentus. Methods Enzymol. 1991, 204, 372–384. [Google Scholar] [PubMed]
- Almqvist, H.; Sandahl, M.; Lidén, G. A rapid method for analysis of fermentatively produced d-xylonate using ultra-high performance liquid chromatography and evaporative light scattering detection. Biosci. Biotechnol. Biochem. 2017, 81, 1078–1080. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Johnsen, U.; Dambeck, M.; Zaiss, H.; Fuhrer, T.; Soppa, J.; Sauer, U.; Schönheit, P. d-xylose degradation pathway in the halophilic archaeon Haloferax volcanii. J. Biol. Chem. 2009, 284, 27290–27303. [Google Scholar] [CrossRef] [PubMed]




| XylB | XylB | XylDXA | |
|---|---|---|---|
| Tris-HCl, pH 8.0 (mM) | 100.0 | 100.0 | 100.0 |
| MgCl2 (mM) | 2.0 | 2.0 | 10.0 |
| NAD+ (mM) | 2.0 | 2.0 | 2.0 |
| Xylose (mM) | 83.3 | - | - |
| Arabinose (mM) | - | 83.3 | - |
| Xylonate (mM) | - | - | 80.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almqvist, H.; Jonsdottir Glaser, S.; Tufvegren, C.; Wasserstrom, L.; Lidén, G. Characterization of the Weimberg Pathway in Caulobacter crescentus. Fermentation 2018, 4, 44. https://doi.org/10.3390/fermentation4020044
Almqvist H, Jonsdottir Glaser S, Tufvegren C, Wasserstrom L, Lidén G. Characterization of the Weimberg Pathway in Caulobacter crescentus. Fermentation. 2018; 4(2):44. https://doi.org/10.3390/fermentation4020044
Chicago/Turabian StyleAlmqvist, Henrik, Sara Jonsdottir Glaser, Celina Tufvegren, Lisa Wasserstrom, and Gunnar Lidén. 2018. "Characterization of the Weimberg Pathway in Caulobacter crescentus" Fermentation 4, no. 2: 44. https://doi.org/10.3390/fermentation4020044
APA StyleAlmqvist, H., Jonsdottir Glaser, S., Tufvegren, C., Wasserstrom, L., & Lidén, G. (2018). Characterization of the Weimberg Pathway in Caulobacter crescentus. Fermentation, 4(2), 44. https://doi.org/10.3390/fermentation4020044